The role of KRAS splice variants in cancer biology
- PMID: 36393833
- PMCID: PMC9663995
- DOI: 10.3389/fcell.2022.1033348
The role of KRAS splice variants in cancer biology
Abstract
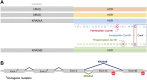
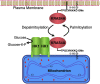
The three mammalian RAS genes (HRAS, NRAS and KRAS) encode four proteins that play central roles in cancer biology. Among them, KRAS is mutated more frequently in human cancer than any other oncogene. The pre-mRNA of KRAS is alternatively spliced to give rise to two products, KRAS4A and KRAS4B, which differ in the membrane targeting sequences at their respective C-termini. Notably, both KRAS4A and KRAS4B are oncogenic when KRAS is constitutively activated by mutation in exon 2 or 3. Whereas KRAS4B is the most studied oncoprotein, KRAS4A is understudied and until recently considered relatively unimportant. Emerging work has confirmed expression of KRAS4A in cancer and found non-overlapping functions of the splice variants. The most clearly demonstrated of these is direct regulation of hexokinase 1 by KRAS4A, suggesting that the metabolic vulnerabilities of KRAS-mutant tumors may be determined in part by the relative expression of the splice variants. The aim of this review is to address the most relevant characteristics and differential functions of the KRAS splice variants as they relate to cancer onset and progression.
Keywords: KRAS; KRAS4A; KRAS4B; alternative splicing; glycolysis; oncogene; oncoprotein.
Copyright © 2022 Nuevo-Tapioles and Philips.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Aran V., Masson Domingues P., Carvalho de Macedo F., Moreira de Sousa C. A., Caldas Montella T., de Souza Accioly M. T., et al. (2018). A cross-sectional study examining the expression of splice variants K-RAS4A and K-RAS4B in advanced non-small-cell lung cancer patients. Lung Cancer 116, 7–14. 10.1016/j.lungcan.2017.12.005 - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

