Automated and Efficient Generation of General Molecular Aggregate Structures
- PMID: 36394430
- PMCID: PMC10107477
- DOI: 10.1002/anie.202214477
Automated and Efficient Generation of General Molecular Aggregate Structures
Abstract
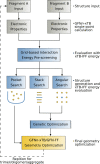
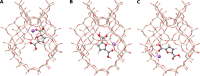
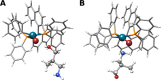
Modeling intermolecular interactions of complex non-covalent structures is important in many areas of chemistry. To facilitate the generation of reasonable dimer, oligomer, and general aggregate geometries, we introduce an automated computational interaction site screening (aISS) workflow. This easy-to-use tool combines a genetic algorithm employing the intermolecular force-field xTB-IFF for initial search steps with the general force-field GFN-FF and the semi-empirical GFN2-xTB method for geometry optimizations. Compared with the alternative CREST program, aISS yields similar results but with computer time savings of 1-3 orders of magnitude. This allows for the treatment of systems with thousands of atoms composed of elements up to radon, e.g., metal-organic complexes, or even polyhedra and zeolite cut-outs which were not accessible before. Moreover, aISS can identify reactive sites and provides options like site-directed (user-guided) screening.
Keywords: Global Optimization; Non-Covalent Interaction; Quantum Chemistry; Supramolecular Chemistry.
© 2022 The Authors. Angewandte Chemie International Edition published by Wiley-VCH GmbH.
Conflict of interest statement
There are no conflicts to declare.
Figures



References
-
- Mahmudov K. T., Kopylovich M. N., Guedes da Silva M. F. C., Pombeiro A. J., Coord. Chem. Rev. 2017, 345, 54.
-
- Zhou Z., Yan X., Cook T. R., Saha M. L., Stang P. J., J. Am. Chem. Soc. 2016, 138, 806. - PubMed
-
- Rest C., Kandanelli R., Fernández G., Chem. Soc. Rev. 2015, 44, 2543. - PubMed
-
- Karshikoff A., Non-Covalent Interactions in Proteins, World Scientific, Singapore, 2021.
-
- Zhou P., Huang J., Tian F., Curr. Med. Chem. 2012, 19, 226. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

