Aorta- and liver-generated TMAO enhances trained immunity for increased inflammation via ER stress/mitochondrial ROS/glycolysis pathways
- PMID: 36394956
- PMCID: PMC9870092
- DOI: 10.1172/jci.insight.158183
Aorta- and liver-generated TMAO enhances trained immunity for increased inflammation via ER stress/mitochondrial ROS/glycolysis pathways
Abstract
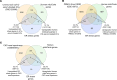
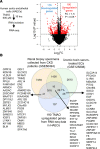
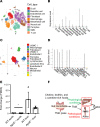
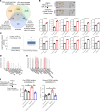
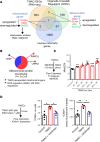
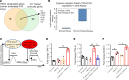
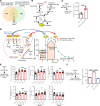
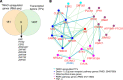
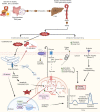
We determined whether gut microbiota-produced trimethylamine (TMA) is oxidized into trimethylamine N-oxide (TMAO) in nonliver tissues and whether TMAO promotes inflammation via trained immunity (TI). We found that endoplasmic reticulum (ER) stress genes were coupregulated with MitoCarta genes in chronic kidney diseases (CKD); TMAO upregulated 190 genes in human aortic endothelial cells (HAECs); TMAO synthesis enzyme flavin-containing monooxygenase 3 (FMO3) was expressed in human and mouse aortas; TMAO transdifferentiated HAECs into innate immune cells; TMAO phosphorylated 12 kinases in cytosol via its receptor PERK and CREB, and integrated with PERK pathways; and PERK inhibitors suppressed TMAO-induced ICAM-1. TMAO upregulated 3 mitochondrial genes, downregulated inflammation inhibitor DARS2, and induced mitoROS, and mitoTEMPO inhibited TMAO-induced ICAM-1. β-Glucan priming, followed by TMAO restimulation, upregulated TNF-α by inducing metabolic reprogramming, and glycolysis inhibitor suppressed TMAO-induced ICAM-1. Our results have provided potentially novel insights regarding TMAO roles in inducing EC activation and innate immune transdifferentiation and inducing metabolic reprogramming and TI for enhanced vascular inflammation, and they have provided new therapeutic targets for treating cardiovascular diseases (CVD), CKD-promoted CVD, inflammation, transplantation, aging, and cancer.
Keywords: Cardiovascular disease; Chronic kidney disease; Immunology; Inflammation; Innate immunity.
Conflict of interest statement
Figures


References
-
- Shao Y, et al. Endothelial immunity trained by coronavirus infections, DAMP stimulations and regulated by anti-oxidant NRF2 may contribute to inflammations, myelopoiesis, COVID-19 cytokine storms and thromboembolism. Front Immunol. 2021;12:653110. doi: 10.3389/fimmu.2021.653110. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK121227/DK/NIDDK NIH HHS/United States
- R01 HL130233/HL/NHLBI NIH HHS/United States
- R01 HL131460/HL/NHLBI NIH HHS/United States
- R01 DK104116/DK/NIDDK NIH HHS/United States
- R01 HL132399/HL/NHLBI NIH HHS/United States
- I01 BX004658/BX/BLRD VA/United States
- R01 DK132888/DK/NIDDK NIH HHS/United States
- K08 HL151747/HL/NHLBI NIH HHS/United States
- R01 HL138749/HL/NHLBI NIH HHS/United States
- R01 HL147565/HL/NHLBI NIH HHS/United States
- R01 DK113775/DK/NIDDK NIH HHS/United States
- R01 DK098511/DK/NIDDK NIH HHS/United States
- R01 HL163570/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

