Treasures from trash in cancer research
- PMID: 36395362
- PMCID: PMC9671455
- DOI: 10.18632/oncotarget.28308
Treasures from trash in cancer research
Abstract
Introduction: Cancer research has significantly improved in recent years, primarily due to next-generation sequencing (NGS) technology. Consequently, an enormous amount of genomic and transcriptomic data has been generated. In most cases, the data needed for research goals are used, and unwanted reads are discarded. However, these eliminated data contain relevant information. Aiming to test this hypothesis, genomic and transcriptomic data were acquired from public datasets.
Materials and methods: Metagenomic tools were used to explore genomic cancer data; additional annotations were used to explore differentially expressed ncRNAs from miRNA experiments, and variants in adjacent to tumor samples from RNA-seq experiments were also investigated.
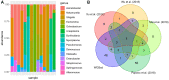
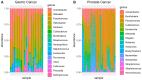
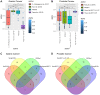
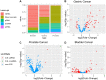
Results: In all analyses, new data were obtained: from DNA-seq data, microbiome taxonomies were characterized with a similar performance of dedicated metagenomic research; from miRNA-seq data, additional differentially expressed sncRNAs were found; and in tumor and adjacent to tumor tissue data, somatic variants were found.
Conclusions: These findings indicate that unexplored data from NGS experiments could help elucidate carcinogenesis and discover putative biomarkers with clinical applications. Further investigations should be considered for experimental design, providing opportunities to optimize data, saving time and resources while granting access to multiple genomic perspectives from the same sample and experimental run.
Keywords: RNA-Seq variant calling; cancer metagenomics; cancer sncRNA expression.
Conflict of interest statement
Authors have no conflicts of interest to declare.
Figures

References
-
- Dash S, Shakyawar SK, Sharma M, Kaushik S. Big data in healthcare: management, analysis and future prospects. Journal of Big Data. 2019; 6:1–25. 10.1186/s40537-019-0217-0 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

