Deep learning for predicting major pathological response to neoadjuvant chemoimmunotherapy in non-small cell lung cancer: A multicentre study
- PMID: 36395737
- PMCID: PMC9672965
- DOI: 10.1016/j.ebiom.2022.104364
Deep learning for predicting major pathological response to neoadjuvant chemoimmunotherapy in non-small cell lung cancer: A multicentre study
Abstract
Background: This study, based on multicentre cohorts, aims to utilize computed tomography (CT) images to construct a deep learning model for predicting major pathological response (MPR) to neoadjuvant chemoimmunotherapy in non-small cell lung cancer (NSCLC) and further explore the biological basis under its prediction.
Methods: 274 patients undergoing curative surgery after neoadjuvant chemoimmunotherapy for NSCLC at 4 centres from January 2019 to December 2021 were included and divided into a training cohort, an internal validation cohort, and an external validation cohort. ShuffleNetV2x05-based features of the primary tumour on the CT scans within the 2 weeks preceding neoadjuvant administration were employed to develop a deep learning score for distinguishing MPR and non-MPR. To reveal the underlying biological basis of the deep learning score, a genetic analysis was conducted based on 25 patients with RNA-sequencing data.
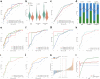
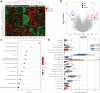
Findings: MPR was achieved in 54.0% (n = 148) patients. The area under the curve (AUC) of the deep learning score to predict MPR was 0.73 (95% confidence interval [CI]: 0.58-0.86) and 0.72 (95% CI: 0.58-0.85) in the internal validation and external validation cohorts, respectively. After integrating the clinical characteristic into the deep learning score, the combined model achieved satisfactory performance in the internal validation (AUC: 0.77, 95% CI: 0.64-0.89) and external validation cohorts (AUC: 0.75, 95% CI: 0.62-0.87). In the biological basis exploration for the deep learning score, a high deep learning score was associated with the downregulation of pathways mediating tumour proliferation and the promotion of antitumour immune cell infiltration in the microenvironment.
Interpretation: The proposed deep learning model could effectively predict MPR in NSCLC patients treated with neoadjuvant chemoimmunotherapy.
Funding: This study was supported by National Key Research and Development Program of China, China (2017YFA0205200); National Natural Science Foundation of China, China (91959126, 82022036, 91959130, 81971776, 81771924, 6202790004, 81930053, 9195910169, 62176013, 8210071009); Beijing Natural Science Foundation, China (L182061); Strategic Priority Research Program of Chinese Academy of Sciences, China (XDB38040200); Chinese Academy of Sciences, China (GJJSTD20170004, QYZDJ-SSW-JSC005); Shanghai Hospital Development Center, China (SHDC2020CR3047B); and Science and Technology Commission of Shanghai Municipality, China (21YF1438200).
Keywords: Deep learning; Major pathological response; Neoadjuvant chemoimmunotherapy; Non-small cell lung cancer.
Copyright © 2022 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests We declare no competing interests.
Figures


References
-
- National Comprehensive Cancer Network. (NCCN) Clinical practice guidelines in oncology. Non-small cell lung cancer, version 1. 2022. https://www.nccn.org/professionals/physician_gls/default.aspx [Internet] - PubMed
-
- Boyd J.A., Hubbs J.L., Kim D.W., Hollis D., Marks L.B., Kelsey C.R. Timing of local and distant failure in resected lung cancer: implications for reported rates of local failure. J Thorac Oncol. 2010;5(2):211–214. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

