GPCRdb in 2023: state-specific structure models using AlphaFold2 and new ligand resources
- PMID: 36395823
- PMCID: PMC9825476
- DOI: 10.1093/nar/gkac1013
GPCRdb in 2023: state-specific structure models using AlphaFold2 and new ligand resources
Abstract
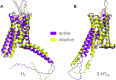
G protein-coupled receptors (GPCRs) are physiologically abundant signaling hubs routing hundreds of extracellular signal substances and drugs into intracellular pathways. The GPCR database, GPCRdb supports >5000 interdisciplinary researchers every month with reference data, analysis, visualization, experiment design and dissemination. Here, we present our fifth major GPCRdb release setting out with an overview of the many resources for receptor sequences, structures, and ligands. This includes recently published additions of class D generic residue numbers, a comparative structure analysis tool to identify functional determinants, trees clustering GPCR structures by 3D conformation, and mutations stabilizing inactive/active states. We provide new state-specific structure models of all human non-olfactory GPCRs built using AlphaFold2-MultiState. We also provide a new resource of endogenous ligands along with a larger number of surrogate ligands with bioactivity, vendor, and physiochemical descriptor data. The one-stop-shop ligand resources integrate ligands/data from the ChEMBL, Guide to Pharmacology, PDSP Ki and PubChem database. The GPCRdb is available at https://gpcrdb.org.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

