Prognostic Prediction of Genotype vs Phenotype in Genetic Cardiomyopathies
- PMID: 36396199
- PMCID: PMC10754019
- DOI: 10.1016/j.jacc.2022.08.804
Prognostic Prediction of Genotype vs Phenotype in Genetic Cardiomyopathies
Abstract
Background: Diverse genetic backgrounds often lead to phenotypic heterogeneity in cardiomyopathies (CMPs). Previous genotype-phenotype studies have primarily focused on the analysis of a single phenotype, and the diagnostic and prognostic features of the CMP genotype across different phenotypic expressions remain poorly understood.
Objectives: We sought to define differences in outcome prediction when stratifying patients based on phenotype at presentation compared with genotype in a large cohort of patients with CMPs and positive genetic testing.
Methods: Dilated cardiomyopathy (DCM), arrhythmogenic right ventricular cardiomyopathy, left-dominant arrhythmogenic cardiomyopathy, and biventricular arrhythmogenic cardiomyopathy were examined in this study. A total of 281 patients (80% DCM) with pathogenic or likely pathogenic variants were included. The primary and secondary outcomes were: 1) all-cause mortality (D)/heart transplant (HT); 2) sudden cardiac death/major ventricular arrhythmias (SCD/MVA); and 3) heart failure-related death (DHF)/HT/left ventricular assist device implantation (LVAD).
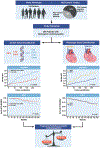
Results: Survival analysis revealed that SCD/MVA events occurred more frequently in patients without a DCM phenotype and in carriers of DSP, PKP2, LMNA, and FLNC variants. However, after adjustment for age and sex, genotype-based classification, but not phenotype-based classification, was predictive of SCD/MVA. LMNA showed the worst trends in terms of D/HT and DHF/HT/LVAD.
Conclusions: Genotypes were associated with significant phenotypic heterogeneity in genetic cardiomyopathies. Nevertheless, in our study, genotypic-based classification showed higher precision in predicting the outcome of patients with CMP than phenotype-based classification. These findings add to our current understanding of inherited CMPs and contribute to the risk stratification of patients with positive genetic testing.
Keywords: ALVC; ARVC; DCM; genotype; pathogenic/likely pathogenic variants; phenotype.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Funding Support and Author Disclosures This study was supported by National Institutes of Health (NIH) grants R01HL69071, R01HL116906, and R01HL147064, and American Heart Association grant 17GRNT33670495 (to Dr Mestroni); NIH grant 1K23HI067915; NIH grants 2UM1HG006542 and R01HL109209 (to Dr Taylor); and CRTrieste Foundation and Cassa di Risparmio di Gorizia Foundation (to Dr Sinagra). This work is also supported by NIH/National Center for Advancing Translational Sciences Colorado CTSA grant numbers UL1 TR002535 and UL1 TR001082. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Figures



Comment in
-
Genotype-Based Risk Stratification Can Outperform Phenotype-Based Practice for Inherited Cardiomyopathies: Not All Paths Are Equal.J Am Coll Cardiol. 2022 Nov 22;80(21):1995-1997. doi: 10.1016/j.jacc.2022.09.018. J Am Coll Cardiol. 2022. PMID: 36396200 No abstract available.
References
-
- Elliott P, Andersson B, Arbustini E, et al. Classification of the cardiomyopathies: a position statement from the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J. 2008;29(2):270–276. - PubMed
-
- Members WC, Ommen SR, Mital S, et al. 2020 AHA/ACC guideline for the diagnosis and treatment of patients with hypertrophic cardiomyopathy. Circulation. 2020;142(25):558–631. - PubMed
-
- Towbin JA, McKenna WJ, Abrams DJ, et al. 2019 HRS expert consensus statement on evaluation, risk stratification, and management of arrhythmogenic cardiomyopathy. Heart Rhythm. 2019;16(11):e301–e372. - PubMed
-
- Golnaz G, Jabbari R, Risgaard B, et al. Mixed phenotypes: implications in family screening of inherited cardiomyopathies. Eur Heart J. 2013;34(Suppl_1), 3772–3772.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

