Structure determination of inactive-state GPCRs with a universal nanobody
- PMID: 36396979
- PMCID: PMC12014012
- DOI: 10.1038/s41594-022-00859-8
Structure determination of inactive-state GPCRs with a universal nanobody
Abstract
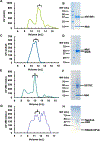
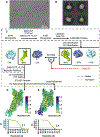
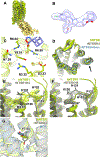
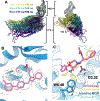
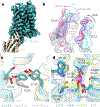
Cryogenic electron microscopy (cryo-EM) has widened the field of structure-based drug discovery by allowing for routine determination of membrane protein structures previously intractable. Despite representing one of the largest classes of therapeutic targets, most inactive-state G protein-coupled receptors (GPCRs) have remained inaccessible for cryo-EM because their small size and membrane-embedded nature impedes projection alignment for high-resolution map reconstructions. Here we demonstrate that the same single-chain camelid antibody (nanobody) recognizing a grafted intracellular loop can be used to obtain cryo-EM structures of inactive-state GPCRs at resolutions comparable or better than those obtained by X-ray crystallography. Using this approach, we obtained structures of neurotensin 1 receptor bound to antagonist SR48692, μ-opioid receptor bound to alvimopan, apo somatostatin receptor 2 and histamine receptor 2 bound to famotidine. We expect this rapid, straightforward approach to facilitate the broad exploration of GPCR inactive states without the need for extensive engineering and crystallization.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures










References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

