Dynamics of Viral and Host 3D Genome Structure upon Infection
- PMID: 36398441
- PMCID: PMC9843816
- DOI: 10.4014/jmb.2208.08020
Dynamics of Viral and Host 3D Genome Structure upon Infection
Abstract
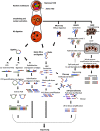
Eukaryotic chromatin is highly organized in the 3D nuclear space and dynamically regulated in response to environmental stimuli. This genomic organization is arranged in a hierarchical fashion to support various cellular functions, including transcriptional regulation of gene expression. Like other host cellular mechanisms, viral pathogens utilize and modulate host chromatin architecture and its regulatory machinery to control features of their life cycle, such as lytic versus latent status. Combined with previous research focusing on individual loci, recent global genomic studies employing conformational assays coupled with high-throughput sequencing technology have informed models for host and, in some cases, viral 3D chromosomal structure re-organization during infection and the contribution of these alterations to virus-mediated diseases. Here, we review recent discoveries and progress in host and viral chromatin structural dynamics during infection, focusing on a subset of DNA (human herpesviruses and HPV) as well as RNA (HIV, influenza virus and SARS-CoV-2) viruses. An understanding of how host and viral genomic structure affect gene expression in both contexts and ultimately viral pathogenesis can facilitate the development of novel therapeutic strategies.
Keywords: Chromatin structure; EBV; HIV; HPV; KSHV; SARS-CoV-2; influenza virus.
Conflict of interest statement
The authors have no financial conflicts of interest to declare.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

