Neutralizing the Impact of the Virulence Factor LecA from Pseudomonas aeruginosa on Human Cells with New Glycomimetic Inhibitors
- PMID: 36398566
- PMCID: PMC10107299
- DOI: 10.1002/anie.202215535
Neutralizing the Impact of the Virulence Factor LecA from Pseudomonas aeruginosa on Human Cells with New Glycomimetic Inhibitors
Abstract
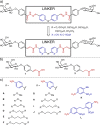
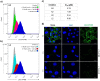
Bacterial adhesion, biofilm formation and host cell invasion of the ESKAPE pathogen Pseudomonas aeruginosa require the tetravalent lectins LecA and LecB, which are therefore drug targets to fight these infections. Recently, we have reported highly potent divalent galactosides as specific LecA inhibitors. However, they suffered from very low solubility and an intrinsic chemical instability due to two acylhydrazone motifs, which precluded further biological evaluation. Here, we isosterically substituted the acylhydrazones and systematically varied linker identity and length between the two galactosides necessary for LecA binding. The optimized divalent LecA ligands showed improved stability and were up to 1000-fold more soluble. Importantly, these properties now enabled their biological characterization. The lead compound L2 potently inhibited LecA binding to lung epithelial cells, restored wound closure in a scratch assay and reduced the invasiveness of P. aeruginosa into host cells.
Keywords: Glycomimetics; Lectin; Pathoblocker; Pseudomonas Aeruginosa; Virulence.
© 2022 The Authors. Angewandte Chemie International Edition published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
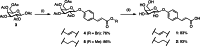






References
-
- Meiers J., Siebs E., Zahorska E., Titz A., Curr. Opin. Chem. Biol. 2019, 53, 51–67. - PubMed
-
- Rice L. B., J. Infect. Dis. 2008, 197, 1079–1081. - PubMed
-
- Wagner S., Sommer R., Hinsberger S., Lu C., Hartmann R. W., Empting M., Titz A., J. Med. Chem. 2016, 59, 5929–5969. - PubMed
-
- Davies D., Nat. Rev. Drug Discovery 2003, 2, 114–122. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

