ATM inhibition drives metabolic adaptation via induction of macropinocytosis
- PMID: 36399181
- PMCID: PMC9679964
- DOI: 10.1083/jcb.202007026
ATM inhibition drives metabolic adaptation via induction of macropinocytosis
Abstract
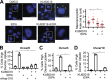
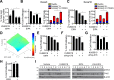
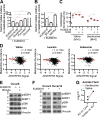
Macropinocytosis is a nonspecific endocytic process that may enhance cancer cell survival under nutrient-poor conditions. Ataxia-Telangiectasia mutated (ATM) is a tumor suppressor that has been previously shown to play a role in cellular metabolic reprogramming. We report that the suppression of ATM increases macropinocytosis to promote cancer cell survival in nutrient-poor conditions. Combined inhibition of ATM and macropinocytosis suppressed proliferation and induced cell death both in vitro and in vivo. Supplementation of ATM-inhibited cells with amino acids, branched-chain amino acids (BCAAs) in particular, abrogated macropinocytosis. Analysis of ATM-inhibited cells in vitro demonstrated increased BCAA uptake, and metabolomics of ascites and interstitial fluid from tumors indicated decreased BCAAs in the microenvironment of ATM-inhibited tumors. These data reveal a novel basis of ATM-mediated tumor suppression whereby loss of ATM stimulates protumorigenic uptake of nutrients in part via macropinocytosis to promote cancer cell survival and reveal a potential metabolic vulnerability of ATM-inhibited cells.
© 2022 Huang et al.
Figures
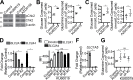




References
-
- Batey, M.A., Zhao Y., Kyle S., Richardson C., Slade A., Martin N.M.B., Lau A., Newell D.R., and Curtin N.J.. 2013. Preclinical evaluation of a novel ATM inhibitor, KU59403, in vitro and in vivo in p53 functional and dysfunctional models of human cancer. Mol. Cancer Ther. 12:959–967. 10.1158/1535-7163.MCT-12-0707 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

