Detecting Liver Cancer Using Cell-Free DNA Fragmentomes
- PMID: 36399356
- PMCID: PMC9975663
- DOI: 10.1158/2159-8290.CD-22-0659
Detecting Liver Cancer Using Cell-Free DNA Fragmentomes
Abstract
Liver cancer is a major cause of cancer mortality worldwide. Screening individuals at high risk, including those with cirrhosis and viral hepatitis, provides an avenue for improved survival, but current screening methods are inadequate. In this study, we used whole-genome cell-free DNA (cfDNA) fragmentome analyses to evaluate 724 individuals from the United States, the European Union, or Hong Kong with hepatocellular carcinoma (HCC) or who were at average or high-risk for HCC. Using a machine learning model that incorporated multifeature fragmentome data, the sensitivity for detecting cancer was 88% in an average-risk population at 98% specificity and 85% among high-risk individuals at 80% specificity. We validated these results in an independent population. cfDNA fragmentation changes reflected genomic and chromatin changes in liver cancer, including from transcription factor binding sites. These findings provide a biological basis for changes in cfDNA fragmentation in patients with liver cancer and provide an accessible approach for noninvasive cancer detection.
Significance: There is a great need for accessible and sensitive screening approaches for HCC worldwide. We have developed an approach for examining genome-wide cfDNA fragmentation features to provide a high-performing and cost-effective approach for liver cancer detection. See related commentary Rolfo and Russo, p. 532. This article is highlighted in the In This Issue feature, p. 517.
©2022 The Authors; Published by the American Association for Cancer Research.
Figures


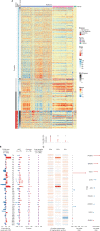

Comment in
-
Moving Forward Liquid Biopsy in Early Liver Cancer Detection.Cancer Discov. 2023 Mar 1;13(3):532-534. doi: 10.1158/2159-8290.CD-22-1439. Cancer Discov. 2023. PMID: 36855918
References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. . Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. - PubMed
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin 2021;71:7–33. - PubMed
-
- Di Bisceglie AM. Hepatitis C and hepatocellular carcinoma. Hepatology 1997;26(3Suppl 1):34S–8S. - PubMed
-
- Donato F, Tagger A, Gelatti U, Parrinello G, Boffetta P, Albertini A, et al. . Alcohol and hepatocellular carcinoma: the effect of lifetime intake and hepatitis virus infections in men and women. Am J Epidemiol 2002;155:323–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

