Deep mutational scans for ACE2 binding, RBD expression, and antibody escape in the SARS-CoV-2 Omicron BA.1 and BA.2 receptor-binding domains
- PMID: 36399443
- PMCID: PMC9674177
- DOI: 10.1371/journal.ppat.1010951
Deep mutational scans for ACE2 binding, RBD expression, and antibody escape in the SARS-CoV-2 Omicron BA.1 and BA.2 receptor-binding domains
Abstract
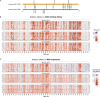
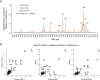
SARS-CoV-2 continues to acquire mutations in the spike receptor-binding domain (RBD) that impact ACE2 receptor binding, folding stability, and antibody recognition. Deep mutational scanning prospectively characterizes the impacts of mutations on these biochemical properties, enabling rapid assessment of new mutations seen during viral surveillance. However, the effects of mutations can change as the virus evolves, requiring updated deep mutational scans. We determined the impacts of all single amino acid mutations in the Omicron BA.1 and BA.2 RBDs on ACE2-binding affinity, RBD folding, and escape from binding by the LY-CoV1404 (bebtelovimab) monoclonal antibody. The effects of some mutations in Omicron RBDs differ from those measured in the ancestral Wuhan-Hu-1 background. These epistatic shifts largely resemble those previously seen in the Alpha variant due to the convergent epistatically modifying N501Y substitution. However, Omicron variants show additional lineage-specific shifts, including examples of the epistatic phenomenon of entrenchment that causes the Q498R and N501Y substitutions present in Omicron to be more favorable in that background than in earlier viral strains. In contrast, the Omicron substitution Q493R exhibits no sign of entrenchment, with the derived state, R493, being as unfavorable for ACE2 binding in Omicron RBDs as in Wuhan-Hu-1. Likely for this reason, the R493Q reversion has occurred in Omicron sub-variants including BA.4/BA.5 and BA.2.75, where the affinity buffer from R493Q reversion may potentiate concurrent antigenic change. Consistent with prior studies, we find that Omicron RBDs have reduced expression, and identify candidate stabilizing mutations that ameliorate this deficit. Last, our maps highlight a broadening of the sites of escape from LY-CoV1404 antibody binding in BA.1 and BA.2 compared to the ancestral Wuhan-Hu-1 background. These BA.1 and BA.2 deep mutational scanning datasets identify shifts in the RBD mutational landscape and inform ongoing efforts in viral surveillance.
Copyright: © 2022 Starr et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: J.D.B. has consulted for Moderna and Merck on viral evolution and epidemiology. T.N.S. and J.D.B. consult for Apriori Bio on deep mutational scanning. T.N.S., A.J.G., and J.D.B. receive a share of intellectual property revenue as inventors on Fred Hutchinson Cancer Center–optioned technology and patents related to deep mutational scanning of viral proteins and stabilization of SARS-CoV-2 RBDs.
Figures


References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

