IMG/VR v4: an expanded database of uncultivated virus genomes within a framework of extensive functional, taxonomic, and ecological metadata
- PMID: 36399502
- PMCID: PMC9825611
- DOI: 10.1093/nar/gkac1037
IMG/VR v4: an expanded database of uncultivated virus genomes within a framework of extensive functional, taxonomic, and ecological metadata
Abstract
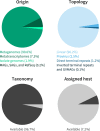
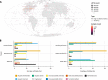
Viruses are widely recognized as critical members of all microbiomes. Metagenomics enables large-scale exploration of the global virosphere, progressively revealing the extensive genomic diversity of viruses on Earth and highlighting the myriad of ways by which viruses impact biological processes. IMG/VR provides access to the largest collection of viral sequences obtained from (meta)genomes, along with functional annotation and rich metadata. A web interface enables users to efficiently browse and search viruses based on genome features and/or sequence similarity. Here, we present the fourth version of IMG/VR, composed of >15 million virus genomes and genome fragments, a ≈6-fold increase in size compared to the previous version. These clustered into 8.7 million viral operational taxonomic units, including 231 408 with at least one high-quality representative. Viral sequences in IMG/VR are now systematically identified from genomes, metagenomes, and metatranscriptomes using a new detection approach (geNomad), and IMG standard annotation are complemented with genome quality estimation using CheckV, taxonomic classification reflecting the latest taxonomic standards, and microbial host taxonomy prediction. IMG/VR v4 is available at https://img.jgi.doe.gov/vr, and the underlying data are available to download at https://genome.jgi.doe.gov/portal/IMG_VR.
© Published by Oxford University Press on behalf of Nucleic Acids Research 2022.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

