Transcriptional reprogramming of infiltrating neutrophils drives lung pathology in severe COVID-19 despite low viral load
- PMID: 36399523
- PMCID: PMC9906672
- DOI: 10.1182/bloodadvances.2022008834
Transcriptional reprogramming of infiltrating neutrophils drives lung pathology in severe COVID-19 despite low viral load
Abstract
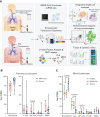
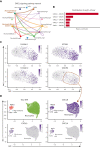
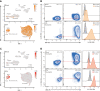
Troubling disparities in COVID-19-associated mortality emerged early, with nearly 70% of deaths confined to Black/African American (AA) patients in some areas. However, targeted studies on this vulnerable population are scarce. Here, we applied multiomics single-cell analyses of immune profiles from matching airways and blood samples of Black/AA patients during acute SARS-CoV-2 infection. Transcriptional reprogramming of infiltrating IFITM2+/S100A12+ mature neutrophils, likely recruited via the IL-8/CXCR2 axis, leads to persistent and self-sustaining pulmonary neutrophilia with advanced features of acute respiratory distress syndrome (ARDS) despite low viral load in the airways. In addition, exacerbated neutrophil production of IL-8, IL-1β, IL-6, and CCL3/4, along with elevated levels of neutrophil elastase and myeloperoxidase, were the hallmarks of transcriptionally active and pathogenic airway neutrophilia. Although our analysis was limited to Black/AA patients and was not designed as a comparative study across different ethnicities, we present an unprecedented in-depth analysis of the immunopathology that leads to acute respiratory distress syndrome in a well-defined patient population disproportionally affected by severe COVID-19.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: F.E.L. is the founder of MicroB-plex, Inc.; serves on the SAB of Be Biopharma Inc.; receives grants from BMGF and Genentech; and receives royalties from BLI, Inc. C.M., D.Y.O., and X.P. are employees of Genentech Inc. D.Y.O. and C.M. own Roche stocks. The remaining authors declare no competing financial interests.
The current affiliation for R.P.R. is Division of Pulmonary, Department of Medicine, Allergy and Critical Care Medicine, University of Pittsburgh School of Medicine, Pittsburgh, PA.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

