Genetic slippage after sex maintains diversity for parasite resistance in a natural host population
- PMID: 36399570
- PMCID: PMC9674289
- DOI: 10.1126/sciadv.abn0051
Genetic slippage after sex maintains diversity for parasite resistance in a natural host population
Abstract
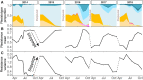
Although parasite-mediated selection is a major driver of host evolution, its influence on genetic variation for parasite resistance is not yet well understood. We monitored resistance in a large population of the planktonic crustacean Daphnia magna over 8 years, as it underwent yearly epidemics of the bacterial pathogen Pasteuria ramosa. We observed cyclic dynamics of resistance: Resistance increased throughout the epidemics, but susceptibility was restored each spring when hosts hatched from sexual resting stages. Host resting stages collected across the year showed that largely resistant host populations can produce susceptible sexual offspring. A genetic model of resistance developed for this host-parasite system, based on multiple loci and strong epistasis, is in partial agreement with our findings. Our results reveal that, despite strong selection for resistance in a natural host population, genetic slippage after sexual reproduction can be a strong factor for the maintenance of genetic diversity of host resistance.
Figures



References
-
- R. A. Fisher, The Genetical Theory of Natural Selection (Oxford Univ. Press, 1930).
-
- Fisher R. A., Polymorphism and natural selection. J. Ecol. 46, 289–293 (1958).
-
- S. A. Frank, Wright’s adaptive landscape versus Fisher’s fundamental theorem, in The Adaptive Landscape in Evolutionary Biology, E. Svensson, R. Calsbeek, Eds. (Oxford Univ. Press, 2013), pp. 41–57.
-
- Jeffery K. J. M., Bangham C. R. M., Do infectious diseases drive MHC diversity? Microbes Infect. 2, 1335–1341 (2000). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

