Unified epigenomic, transcriptomic, proteomic, and metabolomic taxonomy of Alzheimer's disease progression and heterogeneity
- PMID: 36399579
- PMCID: PMC9674284
- DOI: 10.1126/sciadv.abo6764
Unified epigenomic, transcriptomic, proteomic, and metabolomic taxonomy of Alzheimer's disease progression and heterogeneity
Abstract
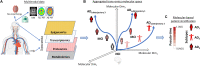
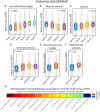
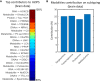
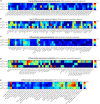
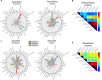
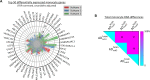
Alzheimer's disease (AD) is a heterogeneous disorder with abnormalities in multiple biological domains. In an advanced machine learning analysis of postmortem brain and in vivo blood multi-omics molecular data (N = 1863), we integrated epigenomic, transcriptomic, proteomic, and metabolomic profiles into a multilevel biological AD taxonomy. We obtained a personalized multilevel molecular index of AD dementia progression that predicts severity of neuropathologies, and identified three robust molecular-based subtypes that explain much of the pathologic and clinical heterogeneity of AD. These subtypes present distinct patterns of alteration in DNA methylation, RNA, proteins, and metabolites, identifiable in the brain and subsequently in blood. In addition, the genetic variations that predispose to the various AD subtypes in brain predict distinct spatial patterns of alteration in cell types, suggesting a unique influence of each putative AD variant on neuropathological mechanisms. These observations support that an individually tailored multi-omics molecular taxonomy of AD may represent distinct targets for preventive or treatment interventions.
Figures



References
-
- Wang B., Mezlini A. M., Demir F., Fiume M., Tu Z., Brudno M., Haibe-Kains B., Goldenberg A., Similarity network fusion for aggregating data types on a genomic scale. Nat. Methods 11, 333–337 (2014). - PubMed
-
- Collisson E. A., Bailey P., Chang D. K., Biankin A. V., Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 16, 207–220 (2019). - PubMed
-
- Hodes R. J., Buckholtz N., Accelerating medicines partnership: Alzheimer’s disease (AMP-AD) knowledge portal aids Alzheimer’s drug discovery through open data sharing. Expert Opin. Ther. Targets 20, 389–391 (2016). - PubMed
-
- Greenwood A. K., Montgomery K. S., Kauer N., Woo K. H., Leanza Z. J., Poehlman W. L., Gockley J., Sieberts S. K., Bradic L., Logsdon B. A., Peters M. A., Omberg L., Mangravite L. M., The AD knowledge portal: A repository for multi-omic data on Alzheimer’s disease and aging. Curr. Protoc. Hum. Genet. 108, e105 (2020). - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

