Biasing the conformation of ELMO2 reveals that myoblast fusion can be exploited to improve muscle regeneration
- PMID: 36400788
- PMCID: PMC9674853
- DOI: 10.1038/s41467-022-34806-4
Biasing the conformation of ELMO2 reveals that myoblast fusion can be exploited to improve muscle regeneration
Abstract
Myoblast fusion is fundamental for the development of multinucleated myofibers. Evolutionarily conserved proteins required for myoblast fusion include RAC1 and its activator DOCK1. In the current study we analyzed the contribution of the DOCK1-interacting ELMO scaffold proteins to myoblast fusion. When Elmo1-/- mice underwent muscle-specific Elmo2 genetic ablation, they exhibited severe myoblast fusion defects. A mutation in the Elmo2 gene that reduced signaling resulted in a decrease in myoblast fusion. Conversely, a mutation in Elmo2 coding for a protein with an open conformation increased myoblast fusion during development and in muscle regeneration. Finally, we showed that the dystrophic features of the Dysferlin-null mice, a model of limb-girdle muscular dystrophy type 2B, were reversed when expressing ELMO2 in an open conformation. These data provide direct evidence that the myoblast fusion process could be exploited for regenerative purposes and improve the outcome of muscle diseases.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
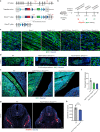
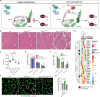
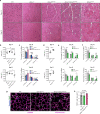
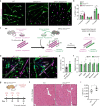
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

