MarineMetagenomeDB: a public repository for curated and standardized metadata for marine metagenomes
- PMID: 36401317
- PMCID: PMC9675116
- DOI: 10.1186/s40793-022-00449-7
MarineMetagenomeDB: a public repository for curated and standardized metadata for marine metagenomes
Abstract
Background: Metagenomics is an expanding field within microbial ecology, microbiology, and related disciplines. The number of metagenomes deposited in major public repositories such as Sequence Read Archive (SRA) and Metagenomic Rapid Annotations using Subsystems Technology (MG-RAST) is rising exponentially. However, data mining and interpretation can be challenging due to mis-annotated and misleading metadata entries. In this study, we describe the Marine Metagenome Metadata Database (MarineMetagenomeDB) to help researchers identify marine metagenomes of interest for re-analysis and meta-analysis. To this end, we have manually curated the associated metadata of several thousands of microbial metagenomes currently deposited at SRA and MG-RAST.
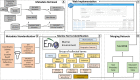
Results: In total, 125 terms were curated according to 17 different classes (e.g., biome, material, oceanic zone, geographic feature and oceanographic phenomena). Other standardized features include sample attributes (e.g., salinity, depth), sample location (e.g., latitude, longitude), and sequencing features (e.g., sequencing platform, sequence count). MarineMetagenomeDB version 1.0 contains 11,449 marine metagenomes from SRA and MG-RAST distributed across all oceans and several seas. Most samples were sequenced using Illumina sequencing technology (84.33%). More than 55% of the samples were collected from the Pacific and the Atlantic Oceans. About 40% of the samples had their biomes assigned as 'ocean'. The 'Quick Search' and 'Advanced Search' tabs allow users to use different filters to select samples of interest dynamically in the web app. The interactive map allows the visualization of samples based on their location on the world map. The web app is also equipped with a novel download tool (on both Windows and Linux operating systems), that allows easy download of raw sequence data of selected samples from their respective repositories. As a use case, we demonstrated how to use the MarineMetagenomeDB web app to select estuarine metagenomes for potential large-scale microbial biogeography studies.
Conclusion: The MarineMetagenomeDB is a powerful resource for non-bioinformaticians to find marine metagenome samples with curated metadata and stimulate meta-studies involving marine microbiomes. Our user-friendly web app is publicly available at https://webapp.ufz.de/marmdb/ .
Keywords: Database; Marine microbiomes; Metadata; Metagenomics; Microbial ecology.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Johnson J, Jain K, Madamwar D. Functional Metagenomics. Curr Dev Biotechnol Bioeng [Internet]. Elsevier; 2017 [cited 2021 Jun 21]. p. 27–43. Available from: https://linkinghub.elsevier.com/retrieve/pii/B978044463667600002X
-
- Qiang-long Z, Shi L, Peng G, Fei-shi L. High-throughput sequencing technology and its application. J Northeast Agric Univ Engl Ed. 2014;21:84–96.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
