Atomistic insights into the binding of SARS-CoV-2 spike receptor binding domain with the human ACE2 receptor: The importance of residue 493
- PMID: 36401897
- PMCID: PMC9595494
- DOI: 10.1016/j.jmgm.2022.108360
Atomistic insights into the binding of SARS-CoV-2 spike receptor binding domain with the human ACE2 receptor: The importance of residue 493
Abstract
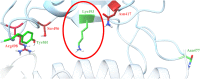
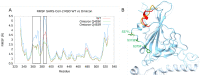
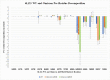
SARS-CoV-2 is a coronavirus that has created a global pandemic. The virus contains a spike protein which has been shown to bind to the ACE2 receptor on the surface of human cells. Vaccines have been developed that recognize elements of the SARS-CoV-2 spike protein and they have been successful in preventing infection. Recently, the Omicron variant of the SARS-CoV-2 virus was reported and quickly became a variant of concern due to its transmissibility. This variant contained an unusually large number (32) of point mutations, of which 15 of those mutations are in the receptor binding domain of the spike protein. While several computational and experimental investigations comparing the binding of the Omicron and wild type RBD to the human ACE2 receptor have been conducted, many of these report contradictory findings. In order to assess the differential binding ability, we conducted 2 μs of classical molecular dynamics (cMD) simulation to estimate the binding affinities and behaviors. Based upon MM-GBSA binding affinity, per-residue energy decomposition analysis, center of mass distance measurements, ensemble clustering, pairwise residue decomposition and hydrogen bonding analysis, our results suggest that a single point mutation is responsible for the enhanced binding of the Omicron mutant relative to the WT. While the 15-point mutations in the receptor binding domain contribute positively and negatively to the affinity of the spike protein for the human ACE2 receptor, it is the point mutation Q493R that confers enhanced binding while the Q493K mutation results in similar binding. The MM-GBSA binding estimations over a 2 μs trajectory, suggest that the wild type binds to ACE2 with a value of -29.69 kcal/mol while the Q493K and Q493R Omicron mutants bind with energy values of -26.67 and -34.56 kcal/mol, respectively. These values are significantly different, given the error estimates associated with the MM-GBSA method. In general, while some mutations increase binding, more mutations diminish binding, leading to an overall similar picture of binding for Q493K and enhanced binding for Q493R.
Keywords: COVID-19; Human ACE2 receptor; MM-GBSA; Molecular dynamics; Omicron; Receptor binding domain; Residue mutations; SARS-CoV-2; Spike protein.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
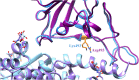

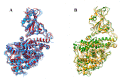
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

