Dynamics and mechanistic interpretations of nonribosomal peptide synthetase cyclization domains
- PMID: 36402006
- PMCID: PMC12173477
- DOI: 10.1016/j.cbpa.2022.102228
Dynamics and mechanistic interpretations of nonribosomal peptide synthetase cyclization domains
Abstract
Ox-/thiazoline groups in nonribosomal peptides are formed by a variant of peptide-forming condensation domains called heterocyclization (Cy) domains and appear in a range of pharmaceutically important natural products and virulence factors. Recent cryo-EM, crystallographic, and NMR studies of Cy domains make it opportune to revisit outstanding questions regarding their molecular mechanisms. This review covers structural and dynamical findings about Cy domains that will inform future bioengineering efforts and our understanding of natural product synthesis.
Keywords: Allostery; Bioengineering; Cyclodehydration; Heterocycle; Pantetheine; Protein–protein interactions.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

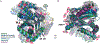
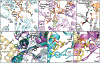

Similar articles
-
Chemical Logic of Peptide Branching by Iterative Nonlinear Nonribosomal Peptide Synthetases.Biochemistry. 2025 Feb 4;64(3):719-734. doi: 10.1021/acs.biochem.4c00749. Epub 2025 Jan 23. Biochemistry. 2025. PMID: 39847710 Review.
-
High-resolution structures of a siderophore-producing cyclization domain from Yersinia pestis offer a refined proposal of substrate binding.J Biol Chem. 2022 Oct;298(10):102454. doi: 10.1016/j.jbc.2022.102454. Epub 2022 Sep 5. J Biol Chem. 2022. PMID: 36063993 Free PMC article.
-
Excision of the epothilone synthetase B cyclization domain and demonstration of in trans condensation/cyclodehydration activity.Biochemistry. 2005 Oct 11;44(40):13385-93. doi: 10.1021/bi051124x. Biochemistry. 2005. PMID: 16201763
-
Diol as a New Pantetheine Surrogate for Chemoenzymatic Synthesis of Cyclic Peptides via Nonribosomal Peptide Cyclases.Methods Mol Biol. 2025;2931:105-121. doi: 10.1007/978-1-0716-4562-8_10. Methods Mol Biol. 2025. PMID: 40531453
-
Structural and functional aspects of the nonribosomal peptide synthetase condensation domain superfamily: discovery, dissection and diversity.Biochim Biophys Acta Proteins Proteom. 2017 Nov;1865(11 Pt B):1587-1604. doi: 10.1016/j.bbapap.2017.05.010. Epub 2017 May 16. Biochim Biophys Acta Proteins Proteom. 2017. PMID: 28526268 Review.
Cited by
-
Structural advances toward understanding the catalytic activity and conformational dynamics of modular nonribosomal peptide synthetases.Nat Prod Rep. 2023 Sep 20;40(9):1550-1582. doi: 10.1039/d3np00003f. Nat Prod Rep. 2023. PMID: 37114973 Free PMC article. Review.
-
The structural basis of substrate selectivity of the acinetobactin biosynthetic adenylation domain, BasE.J Biol Chem. 2025 Apr;301(4):108413. doi: 10.1016/j.jbc.2025.108413. Epub 2025 Mar 15. J Biol Chem. 2025. PMID: 40096888 Free PMC article.
References
-
- Shen Q, Zhou H, Dai G, Zhong G, Huo L, Li A, Liu Y, Yang M, Ravichandran V, Zheng Z, Tang Y-J, Jiao N, Zhang Y, Bian X Characterization of a cryptic NRPS gene cluster in Bacillus velezensis FZB42 reveals a discrete oxidase involved in multithiazole biosynthesis, ACS Catalysis 12 (2022) 3371–3381. doi: 10.1021/acscatal.1c05131. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

