Design and characterization of a high-resolution multiple-SNP capture array by target sequencing for sheep
- PMID: 36402741
- PMCID: PMC9833038
- DOI: 10.1093/jas/skac383
Design and characterization of a high-resolution multiple-SNP capture array by target sequencing for sheep
Abstract
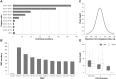
The efficiency of molecular breeding largely depends on inexpensive genotyping arrays. In this study, we aimed to develop an ovine high-resolution multiple-single-nucleotide polymorphism (SNP) capture array, based on genotyping by target sequencing (GBTS) system with capture-in-solution (liquid chip) technology. All the markers were from 40K captured regions, including genes located within selective sweep regions, breed-specific regions, quantitative trait loci (QTL), and the potential functional SNPs on the sheep genome. The results showed that a total of 210K high-quality SNPs were identified in the 40K regions, indicating a high average capture ratio (99.7%) for the target genomic regions. Using genotyped data (n = 317) from liquid chip technology, we further performed genome-wide association studies (GWAS) to detect the genetic loci affecting sheep hair types and teat number. A single significant association signal for hair types was identified on 6.7-7.1 Mb of chromosome 25. The IRF2BP2 gene (chr25: 7,067,974-7,071,785), which is located within this genomic region, has been previously known to be involved in hair/wool traits in sheep. The results further showed a new candidate region around 26.4 Mb of chromosome 13, between the ARHGAP21 and KIAA1217 genes, that was significantly related to teat number in sheep. The haplotype patterns of this region also showed differences in animals with 2, 3, or 4 teats. Advances in using the high-accuracy and low-cost liquid chip are expected to accelerate sheep genomic and breeding studies in the coming years.
Keywords: GWAS; chip design; genotyping by target sequencing; sheep breeding.
Plain language summary
Large-scale genotyping platforms are valuable tools for animal selection and breeding programs. The bead chip has been widely used in both research and commercial applications for a long time. A highly efficient and economical genotyping platform has been developed recently. In the present study, by combining the advantages of resequencing and bead chips, we developed a high-resolution capture array based on target sequencing with capture-in-solution technology (liquid chip), including updated functional probes according to the latest research. We further evaluated this approach by using 317 individuals and found that 210K single-nucleotide polymorphisms can be accurately genotyped, confirming the ratio of the captured regions compared with the designed rations is around 99.7%. Genome-wide association studies conducted using this chip suggested IRF2BP2 gene may be involved in hair types and ARHGAP21-KIAA1217 locus may be related to teats number. The liquid chip with high accuracy and low cost can be widely used in genome-wide association studies and genome selection, supporting efforts in molecular breeding and genetic improvement of sheep.
© The Author(s) 2022. Published by Oxford University Press on behalf of the American Society of Animal Science.
Conflict of interest statement
The authors declare no real or perceived conflicts of interest.
Figures


Similar articles
-
Development and Validation of a 54K Genome-Wide Liquid SNP Chip Panel by Target Sequencing for Dairy Goat.Genes (Basel). 2023 May 22;14(5):1122. doi: 10.3390/genes14051122. Genes (Basel). 2023. PMID: 37239482 Free PMC article.
-
Detection of QTL for greasy fleece weight in sheep using a 50 K single nucleotide polymorphism chip.Trop Anim Health Prod. 2017 Dec;49(8):1657-1662. doi: 10.1007/s11250-017-1373-x. Epub 2017 Aug 11. Trop Anim Health Prod. 2017. PMID: 28801813
-
Development of high-resolution multiple-SNP arrays for genetic analyses and molecular breeding through genotyping by target sequencing and liquid chip.Plant Commun. 2021 Aug 9;2(6):100230. doi: 10.1016/j.xplc.2021.100230. eCollection 2021 Nov 8. Plant Commun. 2021. PMID: 34778746 Free PMC article.
-
Application and prospect of gene chip in genetic breeding of livestock and poultry.Yi Chuan. 2023 Dec 20;45(12):1114-1127. doi: 10.16288/j.yczz.23-233. Yi Chuan. 2023. PMID: 38764275 Review.
-
Genome-wide association studies: an intuitive solution for SNP identification and gene mapping in trees.Funct Integr Genomics. 2023 Sep 12;23(4):297. doi: 10.1007/s10142-023-01224-8. Funct Integr Genomics. 2023. PMID: 37700096 Review.
Cited by
-
Whole Genome Resequencing Identifies Single-Nucleotide Polymorphism Markers of Growth and Reproduction Traits in Zhedong and Zi Crossbred Geese.Genes (Basel). 2023 Feb 14;14(2):487. doi: 10.3390/genes14020487. Genes (Basel). 2023. PMID: 36833414 Free PMC article.
-
Long divergent haplotypes introgressed from wild sheep are associated with distinct morphological and adaptive characteristics in domestic sheep.PLoS Genet. 2023 Feb 23;19(2):e1010615. doi: 10.1371/journal.pgen.1010615. eCollection 2023 Feb. PLoS Genet. 2023. PMID: 36821549 Free PMC article.
-
Establishment of a set of St-group wheat-Thinopyrum ponticum derivative lines conferring resistance to powdery mildew.Front Plant Sci. 2025 Apr 16;16:1576050. doi: 10.3389/fpls.2025.1576050. eCollection 2025. Front Plant Sci. 2025. PMID: 40313730 Free PMC article.
-
Development and validation of a 5K low-density SNP chip for Hainan cattle.BMC Genomics. 2024 Sep 18;25(1):873. doi: 10.1186/s12864-024-10753-w. BMC Genomics. 2024. PMID: 39294563 Free PMC article.
-
Development and Application of a 40 K Liquid Capture Chip for Beef Cattle.Animals (Basel). 2025 May 7;15(9):1346. doi: 10.3390/ani15091346. Animals (Basel). 2025. PMID: 40362161 Free PMC article.
References
-
- Auwera, G. A., Carneiro M. O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., . et al.. 2013. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics 43:11.10.1–11.19.33. doi:10.1002/0471250953.bi1110s43 - DOI - PMC - PubMed
-
- Calvo, J. H., Dervishi E., Sarto P., González-Calvo L., Berzal-Herranz B., Molino F., Serrano M., and Joy M.. . 2013. Structural and functional characterisation of the αS1-casein (CSN1S1) gene and association studies with milk traits in Assaf sheep breed. Livest. Sci. 157:1–8. doi:10.1016/j.livsci.2013.06.014 - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

