How Carvedilol activates β2-adrenoceptors
- PMID: 36402762
- PMCID: PMC9675828
- DOI: 10.1038/s41467-022-34765-w
How Carvedilol activates β2-adrenoceptors
Abstract
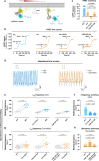
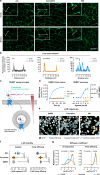
Carvedilol is among the most effective β-blockers for improving survival after myocardial infarction. Yet the mechanisms by which carvedilol achieves this superior clinical profile are still unclear. Beyond blockade of β1-adrenoceptors, arrestin-biased signalling via β2-adrenoceptors is a molecular mechanism proposed to explain the survival benefits. Here, we offer an alternative mechanism to rationalize carvedilol's cellular signalling. Using primary and immortalized cells genome-edited by CRISPR/Cas9 to lack either G proteins or arrestins; and combining biological, biochemical, and signalling assays with molecular dynamics simulations, we demonstrate that G proteins drive all detectable carvedilol signalling through β2ARs. Because a clear understanding of how drugs act is imperative to data interpretation in basic and clinical research, to the stratification of clinical trials or to the monitoring of drug effects on the target pathway, the mechanistic insight gained here provides a foundation for the rational development of signalling prototypes that target the β-adrenoceptor system.
© 2022. The Author(s).
Conflict of interest statement
S.S. is the founder and scientific advisor of 7TM Antibodies GmbH, Jena, Germany. The remaining authors declare no competing interests.
Figures




Comment in
-
How carvedilol does not activate β2-adrenoceptors.Nat Commun. 2023 Nov 30;14(1):7866. doi: 10.1038/s41467-023-42848-5. Nat Commun. 2023. PMID: 38036531 Free PMC article. No abstract available.
References
-
- Nature Medicine Editorial. Mechanism matters. Nat. Med.16, 347 (2010). - PubMed
-
- Mancia G, et al. 2013 ESH/ESC guidelines for the management of arterial hypertension: the Task Force for the Management of Arterial Hypertension of the European Society of Hypertension (ESH) and of the European Society of Cardiology (ESC) Eur. Heart J. 2013;34:2159–2219. doi: 10.1093/eurheartj/eht151. - DOI - PubMed
-
- Martinez-Milla J, Raposeiras-Roubin S, Pascual-Figal DA, Ibanez B. Role of beta-blockers in cardiovascular disease in 2019. Rev. Esp. Cardiol. (Engl. Ed.) 2019;72:844–852. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

