Network location and clustering of genetic mutations determine chronicity in a stylized model of genetic diseases
- PMID: 36402799
- PMCID: PMC9675813
- DOI: 10.1038/s41598-022-23775-9
Network location and clustering of genetic mutations determine chronicity in a stylized model of genetic diseases
Abstract
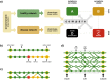
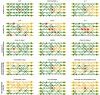
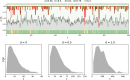
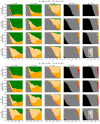
In a highly simplified view, a disease can be seen as the phenotype emerging from the interplay of genetic predisposition and fluctuating environmental stimuli. We formalize this situation in a minimal model, where a network (representing cellular regulation) serves as an interface between an input layer (representing environment) and an output layer (representing functional phenotype). Genetic predisposition for a disease is represented as a loss of function of some network nodes. Reduced, but non-zero, output indicates disease. The simplicity of this genetic disease model and its deep relationship to percolation theory allows us to understand the interplay between disease, network topology and the location and clusters of affected network nodes. We find that our model generates two different characteristics of diseases, which can be interpreted as chronic and acute diseases. In its stylized form, our model provides a new view on the relationship between genetic mutations and the type and severity of a disease.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Network properties of genes harboring inherited disease mutations.Proc Natl Acad Sci U S A. 2008 Mar 18;105(11):4323-8. doi: 10.1073/pnas.0701722105. Epub 2008 Mar 7. Proc Natl Acad Sci U S A. 2008. PMID: 18326631 Free PMC article.
-
Design of double fuzzy clustering-driven context neural networks.Neural Netw. 2018 Aug;104:1-14. doi: 10.1016/j.neunet.2018.03.018. Epub 2018 Apr 9. Neural Netw. 2018. PMID: 29689457
-
Unsupervised deep learning reveals prognostically relevant subtypes of glioblastoma.BMC Bioinformatics. 2017 Oct 3;18(Suppl 11):381. doi: 10.1186/s12859-017-1798-2. BMC Bioinformatics. 2017. PMID: 28984190 Free PMC article.
-
Edgotype: a fundamental link between genotype and phenotype.Curr Opin Genet Dev. 2013 Dec;23(6):649-57. doi: 10.1016/j.gde.2013.11.002. Epub 2013 Nov 26. Curr Opin Genet Dev. 2013. PMID: 24287335 Free PMC article. Review.
-
Ionizing radiation and genetic risks. X. The potential "disease phenotypes" of radiation-induced genetic damage in humans: perspectives from human molecular biology and radiation genetics.Mutat Res. 1999 Aug 11;429(1):45-83. doi: 10.1016/s0027-5107(99)00100-1. Mutat Res. 1999. PMID: 10434024 Review.
References
-
- Cooper R. Disease. Stud. Hist. Philos. Sci. C. 2002;33:263–282.
-
- Ereshefsky M. Defining ‘health’ and ‘disease’. Stud. Hist. Philos. Sci. C. 2009;40:221–227. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

