The non-catalytic role of DNA polymerase epsilon in replication initiation in human cells
- PMID: 36402816
- PMCID: PMC9675812
- DOI: 10.1038/s41467-022-34911-4
The non-catalytic role of DNA polymerase epsilon in replication initiation in human cells
Abstract
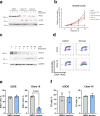
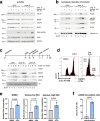
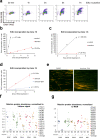
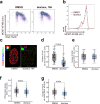
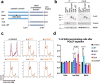
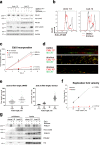
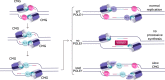
DNA polymerase epsilon (PolE) in an enzyme essential for DNA replication. Deficiencies and mutations in PolE cause severe developmental abnormalities and cancers. Paradoxically, the catalytic domain of yeast PolE catalytic subunit is dispensable for survival, and its non-catalytic essential function is linked with replicative helicase (CMG) assembly. Less is known about the PolE role in replication initiation in human cells. Here we use an auxin-inducible degron system to study the effect of POLE1 depletion on replication initiation in U2OS cells. POLE1-depleted cells were able to assemble CMG helicase and initiate DNA synthesis that failed shortly after. Expression of POLE1 non-catalytic domain rescued this defect resulting in slow, but continuous DNA synthesis. We propose a model where in human U2OS cells POLE1/POLE2 are dispensable for CMG assembly, but essential during later steps of replication initiation. Our study provides some insights into the role of PolE in replication initiation in human cells.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

