Structure of the human heparan sulfate polymerase complex EXT1-EXT2
- PMID: 36402845
- PMCID: PMC9675754
- DOI: 10.1038/s41467-022-34882-6
Structure of the human heparan sulfate polymerase complex EXT1-EXT2
Abstract
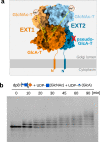
Heparan sulfates are complex polysaccharides that mediate the interaction with a broad range of protein ligands at the cell surface. A key step in heparan sulfate biosynthesis is catalyzed by the bi-functional glycosyltransferases EXT1 and EXT2, which generate the glycan backbone consisting of repeating N-acetylglucosamine and glucuronic acid units. The molecular mechanism of heparan sulfate chain polymerization remains, however, unknown. Here, we present the cryo-electron microscopy structure of human EXT1-EXT2, which reveals the formation of a tightly packed hetero-dimeric complex harboring four glycosyltransferase domains. A combination of in vitro and in cellulo mutational studies is used to dissect the functional role of the four catalytic sites. While EXT1 can catalyze both glycosyltransferase reactions, our results indicate that EXT2 might only have N-acetylglucosamine transferase activity. Our findings provide mechanistic insight into heparan sulfate chain elongation as a nonprocessive process and lay the foundation for future studies on EXT1-EXT2 function in health and disease.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

