Impaired macroglial development and axonal conductivity contributes to the neuropathology of DYRK1A-related intellectual disability syndrome
- PMID: 36402907
- PMCID: PMC9675854
- DOI: 10.1038/s41598-022-24284-5
Impaired macroglial development and axonal conductivity contributes to the neuropathology of DYRK1A-related intellectual disability syndrome
Abstract
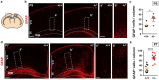
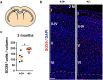
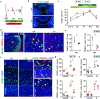
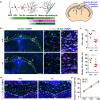
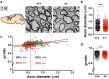
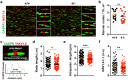
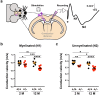
The correct development and activity of neurons and glial cells is necessary to establish proper brain connectivity. DYRK1A encodes a protein kinase involved in the neuropathology associated with Down syndrome that influences neurogenesis and the morphological differentiation of neurons. DYRK1A loss-of-function mutations in heterozygosity cause a well-recognizable syndrome of intellectual disability and autism spectrum disorder. In this study, we analysed the developmental trajectories of macroglial cells and the properties of the corpus callosum, the major white matter tract of the brain, in Dyrk1a+/- mice, a mouse model that recapitulates the main neurological features of DYRK1A syndrome. We found that Dyrk1a+/- haploinsufficient mutants present an increase in astrogliogenesis in the neocortex and a delay in the production of cortical oligodendrocyte progenitor cells and their progression along the oligodendroglial lineage. There were fewer myelinated axons in the corpus callosum of Dyrk1a+/- mice, axons that are thinner and with abnormal nodes of Ranvier. Moreover, action potential propagation along myelinated and unmyelinated callosal axons was slower in Dyrk1a+/- mutants. All these alterations are likely to affect neuronal circuit development and alter network synchronicity, influencing higher brain functions. These alterations highlight the relevance of glial cell abnormalities in neurodevelopmental disorders.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

