H3AGWAS: a portable workflow for genome wide association studies
- PMID: 36402955
- PMCID: PMC9675212
- DOI: 10.1186/s12859-022-05034-w
H3AGWAS: a portable workflow for genome wide association studies
Abstract
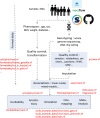
Background: Genome-wide association studies (GWAS) are a powerful method to detect associations between variants and phenotypes. A GWAS requires several complex computations with large data sets, and many steps may need to be repeated with varying parameters. Manual running of these analyses can be tedious, error-prone and hard to reproduce.
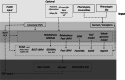
Results: The H3AGWAS workflow from the Pan-African Bioinformatics Network for H3Africa is a powerful, scalable and portable workflow implementing pre-association analysis, implementation of various association testing methods and post-association analysis of results.
Conclusions: The workflow is scalable-laptop to cluster to cloud (e.g., SLURM, AWS Batch, Azure). All required software is containerised and can run under Docker or Singularity.
Keywords: Association testing; Docker; Genome-wide association study; Nextflow; Pipeline; Post-association analysis; Quality control; Singularity; Workflow.
© 2022. The Author(s).
Conflict of interest statement
There is no conflict of interest.
There is no conflict of interest.
Figures
References
-
- Uffelmann E, Huang QQ, Munung NS, de Vries J, Okada Y, Martin AR, et al. Genome-wide association studies. Nat Rev Methods Primers. 2021;1(1):1–21.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

