The envelope proteins from SARS-CoV-2 and SARS-CoV potently reduce the infectivity of human immunodeficiency virus type 1 (HIV-1)
- PMID: 36403071
- PMCID: PMC9675205
- DOI: 10.1186/s12977-022-00611-6
The envelope proteins from SARS-CoV-2 and SARS-CoV potently reduce the infectivity of human immunodeficiency virus type 1 (HIV-1)
Abstract
Background: Viroporins are virally encoded ion channels involved in virus assembly and release. Human immunodeficiency virus type 1 (HIV-1) and influenza A virus encode for viroporins. The human coronavirus SARS-CoV-2 encodes for at least two viroporins, a small 75 amino acid transmembrane protein known as the envelope (E) protein and a larger 275 amino acid protein known as Orf3a. Here, we compared the replication of HIV-1 in the presence of four different β-coronavirus E proteins.
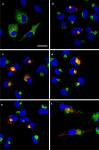
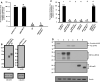
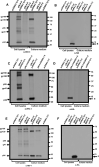
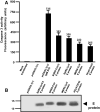
Results: We observed that the SARS-CoV-2 and SARS-CoV E proteins reduced the release of infectious HIV-1 yields by approximately 100-fold while MERS-CoV or HCoV-OC43 E proteins restricted HIV-1 infectivity to a lesser extent. Mechanistically, neither reverse transcription nor mRNA synthesis was involved in the restriction. We also show that all four E proteins caused phosphorylation of eIF2-α at similar levels and that lipidation of LC3-I could not account for the differences in restriction. However, the level of caspase 3 activity in transfected cells correlated with HIV-1 restriction in cells. Finally, we show that unlike the Vpu protein of HIV-1, the four E proteins did not significantly down-regulate bone marrow stromal cell antigen 2 (BST-2).
Conclusions: The results of this study indicate that while viroporins from homologous viruses can enhance virus release, we show that a viroporin from a heterologous virus can suppress HIV-1 protein synthesis and release of infectious virus.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Update of
-
The Envelope Proteins from SARS-CoV-2 and SARS-CoV Potently Reduce the Infectivity of Human Immunodeficiency Virus type 1 (HIV-1).Res Sq [Preprint]. 2022 Oct 24:rs.3.rs-2175808. doi: 10.21203/rs.3.rs-2175808/v1. Res Sq. 2022. Update in: Retrovirology. 2022 Nov 19;19(1):25. doi: 10.1186/s12977-022-00611-6. PMID: 36324807 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

