COVID-19 diagnostics: Molecular biology to nanomaterials
- PMID: 36403665
- PMCID: PMC9673061
- DOI: 10.1016/j.cca.2022.11.017
COVID-19 diagnostics: Molecular biology to nanomaterials
Abstract
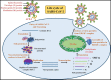
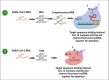
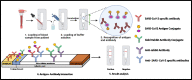
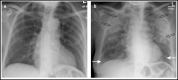
The SARS-CoV-2 pandemic has claimed around 6.4 million lives worldwide. The disease symptoms range from mild flu-like infection to life-threatening complications. The widespread infection demands rapid, simple, and accurate diagnosis. Currently used methods include molecular biology-based approaches that consist of conventional amplification by RT-PCR, isothermal amplification-based techniques such as RT-LAMP, and gene editing tools like CRISPR-Cas. Other methods include immunological detection including ELISA, lateral flow immunoassay, chemiluminescence, etc. Radiological-based approaches are also being used. Despite good analytical performance of these current methods, there is an unmet need for less costly and simpler tests that may be performed at point of care. Accordingly, nanomaterial-based testing has been extensively pursued. In this review, we discuss the currently used diagnostic techniques for SARS-CoV-2, their usefulness, and limitations. In addition, nanoparticle-based approaches have been highlighted as another potential means of detection. The review provides a deep insight into the current diagnostic methods and future trends to combat this deadly menace.
Keywords: Diagnosis; Immunological detection; Molecular detection; Nanoparticles; Radiological approach; SARS-CoV-2.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- WHO Coronavirus (COVID-19) Dashboard, 2021. https://covid19.who.int/.
-
- M. Cascella, M. Rajnik, A. Aleem, S.C. Dulebohn, R.D. Napoli, Features, Evaluation, and Treatment of Coronavirus (COVID-19), (2021) 21. - PubMed
-
- Science Brief: Emerging SARS-CoV-2 Variants, 2021. https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scienti....
Further reading
-
- Dundon W.G., Settypalli T.B.K., Spiegel K., Steinrigl A., Revilla-Fernández S., Schmoll F., Naletoski I., Lamien C.E., Cattoli G. Comparison of eleven in vitro diagnostic assays for the detection of SARS-CoV-2 RNA. J. Virol. Methods. 2021;295 doi: 10.1016/j.jviromet.2021.114200. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

