Biomarkers for isolated congenital heart disease based on maternal amniotic fluid metabolomics analysis
- PMID: 36404327
- PMCID: PMC9677635
- DOI: 10.1186/s12872-022-02912-2
Biomarkers for isolated congenital heart disease based on maternal amniotic fluid metabolomics analysis
Abstract
Introduction: Congenital heart disease (CHD) is one of the most prevalent birth defects in the world. The pathogenesis of CHD is complex and unclear. With the development of metabolomics technology, variations in metabolites may provide new clues about the causes of CHD and may serve as a biomarker during pregnancy.
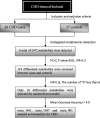
Methods: Sixty-five amniotic fluid samples (28 cases and 37 controls) during the second and third trimesters were utilized in this study. The metabolomics of CHD and normal fetuses were analyzed by untargeted metabolomics technology. Differential comparison and randomForest were used to screen metabolic biomarkers.
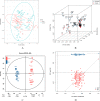
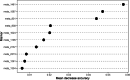
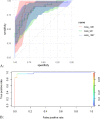
Results: A total of 2472 metabolites were detected, and they were distributed differentially between the cases and controls. Setting the selection criteria of fold change (FC) ≥ 2, P value < 0.01 and variable importance for the projection (VIP) ≥ 1.5, we screened 118 differential metabolites. Within the prediction model by random forest, PE(MonoMe(11,5)/MonoMe(13,5)), N-feruloylserotonin and 2,6-di-tert-butylbenzoquinone showed good prediction effects. Differential metabolites were mainly concentrated in aldosterone synthesis and secretion, drug metabolism, nicotinate and nicotinamide metabolism pathways, which may be related to the occurrence and development of CHD.
Conclusion: This study provides a new database of CHD metabolic biomarkers and mechanistic research. These results need to be further verified in larger samples.
Keywords: Biomarker; Congenital heart disease; Diagnosis; Metabolomics; Reproduction.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no actual or potential competing interests.
Figures

Similar articles
-
Amniotic fluid microbiota and metabolism with non-syndromic congenital heart defects: a multi-omics analysis.BMC Pregnancy Childbirth. 2025 Feb 8;25(1):130. doi: 10.1186/s12884-025-07218-7. BMC Pregnancy Childbirth. 2025. PMID: 39922995 Free PMC article.
-
Biomarkers for congenital ventricular outflow tract malformations based on maternal serum lipid metabolomics analysis.BMC Pregnancy Childbirth. 2024 Aug 20;24(1):547. doi: 10.1186/s12884-024-06738-y. BMC Pregnancy Childbirth. 2024. PMID: 39164614 Free PMC article.
-
Study on the Potential Biomarkers of Maternal Urine Metabolomics for Fetus with Congenital Heart Diseases Based on Modified Gas Chromatograph-Mass Spectrometer.Biomed Res Int. 2019 May 6;2019:1905416. doi: 10.1155/2019/1905416. eCollection 2019. Biomed Res Int. 2019. PMID: 31198782 Free PMC article.
-
Potential role of "omics" technique in prenatal diagnosis of congenital heart defects.Clin Chim Acta. 2018 Jul;482:185-190. doi: 10.1016/j.cca.2018.04.011. Epub 2018 Apr 10. Clin Chim Acta. 2018. PMID: 29649453 Review.
-
Untargeted metabolomics: an overview of its usefulness and future potential in prenatal diagnosis.Expert Rev Proteomics. 2018 Oct;15(10):809-816. doi: 10.1080/14789450.2018.1526678. Epub 2018 Oct 4. Expert Rev Proteomics. 2018. PMID: 30239246 Review.
Cited by
-
Amniotic fluid microbiota and metabolism with non-syndromic congenital heart defects: a multi-omics analysis.BMC Pregnancy Childbirth. 2025 Feb 8;25(1):130. doi: 10.1186/s12884-025-07218-7. BMC Pregnancy Childbirth. 2025. PMID: 39922995 Free PMC article.
-
Biomarkers for congenital ventricular outflow tract malformations based on maternal serum lipid metabolomics analysis.BMC Pregnancy Childbirth. 2024 Aug 20;24(1):547. doi: 10.1186/s12884-024-06738-y. BMC Pregnancy Childbirth. 2024. PMID: 39164614 Free PMC article.
-
Precision fetal cardiology detects cyanotic congenital heart disease using maternal saliva metabolome and artificial intelligence.Sci Rep. 2025 Jan 15;15(1):2060. doi: 10.1038/s41598-025-85216-7. Sci Rep. 2025. PMID: 39814838 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

