Plasma metabolomics and gene regulatory networks analysis reveal the role of nonstructural SARS-CoV-2 viral proteins in metabolic dysregulation in COVID-19 patients
- PMID: 36404352
- PMCID: PMC9676188
- DOI: 10.1038/s41598-022-24170-0
Plasma metabolomics and gene regulatory networks analysis reveal the role of nonstructural SARS-CoV-2 viral proteins in metabolic dysregulation in COVID-19 patients
Abstract
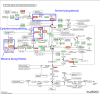
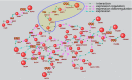
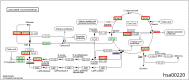
Metabolomic analysis of blood plasma samples from COVID-19 patients is a promising approach allowing for the evaluation of disease progression. We performed the metabolomic analysis of plasma samples of 30 COVID-19 patients and the 19 controls using the high-performance liquid chromatography (HPLC) coupled with tandem mass spectrometric detection (LC-MS/MS). In our analysis, we identified 103 metabolites enriched in KEGG metabolic pathways such as amino acid metabolism and the biosynthesis of aminoacyl-tRNAs, which differed significantly between the COVID-19 patients and the controls. Using ANDSystem software, we performed the reconstruction of gene networks describing the potential genetic regulation of metabolic pathways perturbed in COVID-19 patients by SARS-CoV-2 proteins. The nonstructural proteins of SARS-CoV-2 (orf8 and nsp5) and structural protein E were involved in the greater number of regulatory pathways. The reconstructed gene networks suggest the hypotheses on the molecular mechanisms of virus-host interactions in COVID-19 pathology and provide a basis for the further experimental and computer studies of the regulation of metabolic pathways by SARS-CoV-2 proteins. Our metabolomic analysis suggests the need for nonstructural protein-based vaccines and the control strategy to reduce the disease progression of COVID-19.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- FWNR-2022-0020/Ministry of Science and Higher Education of the Russian Federation
- FSUS-2020-0035/Ministry of Science and Higher Education of the Russian Federation
- 075-15-2021-944/Ministry of Science and Higher Education of the Russian Federation
- FSUS-2020-0035/Ministry of Science and Higher Education of the Russian Federation
- 20-04-60314/Russian Foundation for Basic Research
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

