Alfin-like transcription factor VqAL4 regulates a stilbene synthase to enhance powdery mildew resistance in grapevine
- PMID: 36404575
- PMCID: PMC9831286
- DOI: 10.1111/mpp.13280
Alfin-like transcription factor VqAL4 regulates a stilbene synthase to enhance powdery mildew resistance in grapevine
Abstract
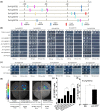
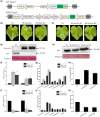
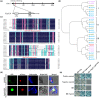
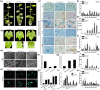
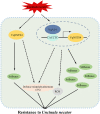
Resveratrol is a phytoalexin that is synthesized by stilbene synthase (STS). Resveratrol in the human diet is known to have beneficial effects on health. We previously identified six novel STS (VqNSTS) transcripts from the transcriptome data of Vitis quinquangularis accession Danfeng-2. However, the functions of and defensive mechanisms triggered by these VqNSTS transcripts remain unknown. In the present study, we demonstrate that the expression of five of these six novel members, VqNSTS2-VqNSTS6, can be induced by the powdery mildew-causing fungus Uncinula necator. Additionally, overexpression of VqNSTS4 in the V. vinifera susceptible cultivar Thompson Seedless promoted accumulation of stilbenes and enhanced resistance to U. necator by activating salicylic acid (SA) signalling. Furthermore, our results indicate that the Alfin-like (AL) transcription factor VqAL4 can directly bind to the G-rich element (CACCTC) in the VqNSTS4 promoter and activate gene expression. Moreover, overexpression of VqAL4 in Thompson Seedless enhanced resistance to U. necator by promoting stilbene accumulation and activating SA signalling. Conversely, RNA interference-mediated silencing of VqNSTS4 and VqAL4 resulted in increased susceptibility to U. necator. Collectively, our results reveal that VqNSTS4, regulated by VqAL4, enhances grapevine resistance to powdery mildew by activating SA signalling. Our findings may be useful to improve disease resistance in perennial fruit trees.
Keywords: Chinese wild Vitis quinquangularis; VqAL4 transcription factor; VqNSTS; disease resistance; stilbene.
© 2022 The Authors. Molecular Plant Pathology published by British Society for Plant Pathology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare they have no conflicts of interest.
Figures




References
-
- Bastola, D.R. , Pethe, V.V. & Winicov, I. (1998) Alfin1, a novel zinc‐finger protein in alfalfa roots that binds to promoter elements in the salt‐inducible MsPRP2 gene. Plant Molecular Biology, 38, 1123–1135. - PubMed
-
- Cao, J. (2012) Characterization of stilbene synthase gene and genetic transformation in grape. Master thesis. Yangling, China: Northwest A & F University.
-
- Chandrika, N.N.P. , Sundaravelpandian, K. , Yu, S.‐M. & Schmidt, W. (2013) ALFIN‐LIKE 6 is involved in root hair elongation during phosphate deficiency in Arabidopsis . New Phytologist, 198, 709–720. - PubMed
-
- Cheng, S. , Xie, X. , Xu, Y. , Zhang, C. , Wang, X. , Zhang, J. et al. (2016) Genetic transformation of a fruit‐specific, highly expressed stilbene synthase gene from Chinese wild Vitis quinquangularis . Planta, 243, 1041–1053. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

