GPI-anchored ligand-BioID2-tagging system identifies Galectin-1 mediating Zika virus entry
- PMID: 36404916
- PMCID: PMC9668739
- DOI: 10.1016/j.isci.2022.105481
GPI-anchored ligand-BioID2-tagging system identifies Galectin-1 mediating Zika virus entry
Abstract
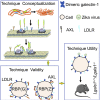
Identification of host factors facilitating pathogen entry is critical for preventing infectious diseases. Here, we report a tagging system consisting of a viral receptor-binding protein (RBP) linked to BioID2, which is expressed on the cell surface via a GPI anchor. Using VSV or Zika virus (ZIKV) RBP, the system (BioID2- RBP(V)-GPI; BioID2-RBP(Z)-GPI) faithfully identifies LDLR and AXL, the receptors of VSV and ZIKV, respectively. Being GPI-anchored is essential for the probe to function properly. Furthermore, BioID2-RBP(Z)-GPI expressed in human neuronal progenitor cells identifies galectin-1 on cell surface pivotal for ZIKV entry. This conclusion is further supported by antibody blocking and galectin-1 silencing in A549 and mouse neural cells. Importantly, Lgals1 -/- mice are significantly more resistant to ZIKV infection than Lgals1 +/+ littermates are, having significantly lower virus titers and fewer pathologies in various organs. This tagging system may have broad applications for identifying protein-protein interactions on the cell surface.
Keywords: Biological sciences; Microbiology; Molecular biology; Virology.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Loss of the TAM Receptor Axl Ameliorates Severe Zika Virus Pathogenesis and Reduces Apoptosis in Microglia.iScience. 2019 Mar 29;13:339-350. doi: 10.1016/j.isci.2019.03.003. Epub 2019 Mar 5. iScience. 2019. PMID: 30884311 Free PMC article.
-
β-Catenin Restricts Zika Virus Internalization by Downregulating Axl.J Virol. 2021 Aug 10;95(17):e0070521. doi: 10.1128/JVI.00705-21. Epub 2021 Aug 10. J Virol. 2021. PMID: 34260264 Free PMC article.
-
Zika Virus Induces Mitotic Catastrophe in Human Neural Progenitors by Triggering Unscheduled Mitotic Entry in the Presence of DNA Damage While Functionally Depleting Nuclear PNKP.J Virol. 2022 May 11;96(9):e0033322. doi: 10.1128/jvi.00333-22. Epub 2022 Apr 12. J Virol. 2022. PMID: 35412344 Free PMC article.
-
Probing Molecular Insights into Zika Virus⁻Host Interactions.Viruses. 2018 May 2;10(5):233. doi: 10.3390/v10050233. Viruses. 2018. PMID: 29724036 Free PMC article. Review.
-
Hide and Seek: The Interplay Between Zika Virus and the Host Immune Response.Front Immunol. 2021 Oct 21;12:750365. doi: 10.3389/fimmu.2021.750365. eCollection 2021. Front Immunol. 2021. PMID: 34745123 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

