Medication based machine learning to identify subpopulations of pediatric hemodialysis patients in an electronic health record database
- PMID: 36405250
- PMCID: PMC9674326
- DOI: 10.1016/j.imu.2022.101104
Medication based machine learning to identify subpopulations of pediatric hemodialysis patients in an electronic health record database
Abstract
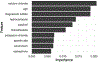
Electronic health records (EHRs) have given rise to large and complex databases of medical information that have the potential to become powerful tools for clinical research. However, differences in coding systems and the detail and accuracy of the information within EHRs can vary across institutions. This makes it challenging to identify subpopulations of patients and limits the widespread use of multi-institutional databases. In this study, we leveraged machine learning to identify patterns in medication usage among hospitalized pediatric patients receiving renal replacement therapy and created a predictive model that successfully differentiated between intermittent (iHD) and continuous renal replacement therapy (CRRT) hemodialysis patients. We trained six machine learning algorithms (logistical regression, Naïve Bayes, k-nearest neighbor, support vector machine, random forest, and gradient boosted trees) using patient records from a multi-center database (n = 533) and prescribed medication ingredients (n = 228) as features to discriminate between the two hemodialysis types. Predictive skill was assessed using a 5-fold cross-validation, and the algorithms showed a range of performance from 0.7 balanced accuracy (logistical regression) to 0.86 (random forest). The two best performing models were further tested using an independent single-center dataset and achieved 84-87% balanced accuracy. This model overcomes issues inherent within large databases and will allow us to utilize and combine historical records, significantly increasing population size and diversity within both iHD and CRRT populations for future clinical studies. Our work demonstrates the utility of using medications alone to accurately differentiate subpopulations of patients in large datasets, allowing codes to be transferred between different coding systems. This framework has the potential to be used to distinguish other subpopulations of patients where discriminatory ICD codes are not available, permitting more detailed insights and new lines of research.
Keywords: Electronic health records; Hemodialysis; Machine learning; Medications; Pediatrics.
Figures


References
-
- Henry J, Pylypchuk Y, Searcy T, Patel V. Adoption of electronic health record systems among U.S. Non-federal acute care hospitals: 2008-2015. In: Office of the National Coordinator for Health Information Technology, editor35. Washington D.C.: ONC Data Brief; 2016.
-
- Roth JA, et al. Introduction to machine learning in digital healthcare epidemiology. Infect Control Hosp Epidemiol 2018;39(12):1457–62. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
