Role of Cancer Stem-like Cells in the Process of Invasion and Mesenchymal Transformation by a Reconstituted Triple-negative Breast Cancer Cell Population Resistant to p53-induced Apoptosis
- PMID: 36405550
- PMCID: PMC9631983
- DOI: 10.1267/ahc.22-00076
Role of Cancer Stem-like Cells in the Process of Invasion and Mesenchymal Transformation by a Reconstituted Triple-negative Breast Cancer Cell Population Resistant to p53-induced Apoptosis
Abstract
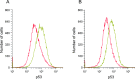
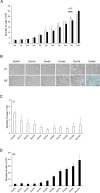
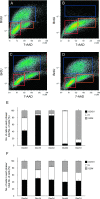
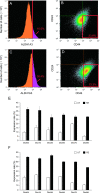
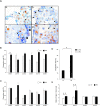
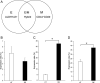
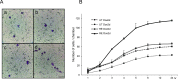
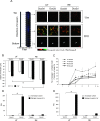
We investigated the role of cancer stem cells (CSCs) in a population of triple-negative breast cancer (TNBC) cells that are resistant to apoptosis. A human breast cancer cell population capable of inducing p53 expression with doxycycline (Dox) was created and used as an untreated control (UT). After the addition of Dox to UT for 5 days, the cell population reconstituted with cells showing resistance to apoptosis was named RE. Fluorescence-activated cell sorting (FACS) and immunostaining revealed that after the addition of Dox, the ratio of cells in the S and G2/M phases decreased in UT as apoptosis proceeded, but did not markedly change in apoptosis-resistant RE. CSC-like cells in RE exhibited a cell morphology with a larger ratio of the major/minor axis than UT. FACS showed that RE had a higher proportion of CSC-like cells and contained more CD44+CD24- mesenchymal CSCs than ALDH1A3+ epithelial-like CSCs. In a Matrigel invasion assay, UT was more likely to form a three-dimensional cell population, whereas RE exhibited a planar population, higher migration ability, and the up-regulated expression of epithelial-mesenchymal transition-related genes. These results provide insights into the mechanisms by which TNBC cells acquire treatment resistance at the time of recurrence.
Keywords: apoptosis resistance; breast cancer; cancer stem-like cell; invasion.
2022 The Japan Society of Histochemistry and Cytochemistry.
Conflict of interest statement
VThe authors declare that there are no conflicts of interest.
Figures
Similar articles
-
Dual inhibition of Wnt and Yes-associated protein signaling retards the growth of triple-negative breast cancer in both mesenchymal and epithelial states.Mol Oncol. 2018 Apr;12(4):423-440. doi: 10.1002/1878-0261.12167. Epub 2018 Feb 21. Mol Oncol. 2018. PMID: 29316250 Free PMC article.
-
Interferon-beta represses cancer stem cell properties in triple-negative breast cancer.Proc Natl Acad Sci U S A. 2017 Dec 26;114(52):13792-13797. doi: 10.1073/pnas.1713728114. Epub 2017 Dec 11. Proc Natl Acad Sci U S A. 2017. PMID: 29229854 Free PMC article.
-
Inhibition of Cdk2 kinase activity selectively targets the CD44⁺/CD24⁻/Low stem-like subpopulation and restores chemosensitivity of SUM149PT triple-negative breast cancer cells.Int J Oncol. 2014 Sep;45(3):1193-9. doi: 10.3892/ijo.2014.2523. Epub 2014 Jun 25. Int J Oncol. 2014. PMID: 24970653 Free PMC article.
-
CSCs in Breast Cancer-One Size Does Not Fit All: Therapeutic Advances in Targeting Heterogeneous Epithelial and Mesenchymal CSCs.Cancers (Basel). 2019 Aug 7;11(8):1128. doi: 10.3390/cancers11081128. Cancers (Basel). 2019. PMID: 31394796 Free PMC article. Review.
-
Breast cancer stem cells and intrinsic subtypes: controversies rage on.Curr Stem Cell Res Ther. 2009 Jan;4(1):50-60. doi: 10.2174/157488809787169110. Curr Stem Cell Res Ther. 2009. PMID: 19149630 Review.
Cited by
-
Cancer stem cell specificity as new targets in breast tumor treatment.Oncol Res. 2025 Mar 19;33(4):811-819. doi: 10.32604/or.2024.050505. eCollection 2025. Oncol Res. 2025. PMID: 40191717 Free PMC article. Review.
-
Immunolocalization of Cytoplasmic ER in ER-negative Breast Carcinoma as a Potent Favorable Prognostic Predictor.Acta Histochem Cytochem. 2023 Aug 30;56(4):59-66. doi: 10.1267/ahc.23-00016. Epub 2023 Aug 23. Acta Histochem Cytochem. 2023. PMID: 37680573 Free PMC article.
References
-
- Aranda, E. and Owen, G. I. (2009) A semi-quantitative assay to screen for angiogenic compounds and compounds with angiogenic potential using the EA.hy926 endothelial cell line. Biol. Res. 42; 377–389. - PubMed
-
- Biddle, A., Liang, X., Gammon, L., Fazil, B., Harper, L. J., Emich, H., et al. (2011) Cancer stem cells in squamous cell carcinoma switch between two distinct phenotypes that are preferentially migratory or proliferative. Cancer Res. 71; 5317–5326. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

