Omicron BA.2 lineage predominance in severe acute respiratory syndrome coronavirus 2 positive cases during the third wave in North India
- PMID: 36405589
- PMCID: PMC9666497
- DOI: 10.3389/fmed.2022.955930
Omicron BA.2 lineage predominance in severe acute respiratory syndrome coronavirus 2 positive cases during the third wave in North India
Abstract
Background: Recent studies on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reveal that Omicron variant BA.1 and sub-lineages have revived the concern over resistance to antiviral drugs and vaccine-induced immunity. The present study aims to analyze the clinical profile and genome characterization of the SARS-CoV-2 variant in eastern Uttar Pradesh (UP), North India.
Methods: Whole-genome sequencing (WGS) was conducted for 146 SARS-CoV-2 samples obtained from individuals who tested coronavirus disease 2019 (COVID-19) positive between the period of 1 January 2022 and 24 February 2022, from three districts of eastern UP. The details regarding clinical and hospitalized status were captured through telephonic interviews after obtaining verbal informed consent. A maximum-likelihood phylogenetic tree was created for evolutionary analysis using MEGA7.
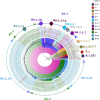
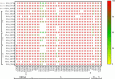
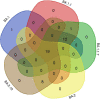
Results: The mean age of study participants was 33.9 ± 13.1 years, with 73.5% accounting for male patients. Of the 98 cases contacted by telephone, 30 (30.6%) had a travel history (domestic/international), 16 (16.3%) reported having been infected with COVID-19 in past, 79 (80.6%) had symptoms, and seven had at least one comorbidity. Most of the sequences belonged to the Omicron variant, with BA.1 (6.2%), BA.1.1 (2.7%), BA.1.1.1 (0.7%), BA.1.1.7 (5.5%), BA.1.17.2 (0.7%), BA.1.18 (0.7%), BA.2 (30.8%), BA.2.10 (50.7%), BA.2.12 (0.7%), and B.1.617.2 (1.3%) lineages. BA.1 and BA.1.1 strains possess signature spike mutations S:A67V, S:T95I, S:R346K, S:S371L, S:G446S, S:G496S, S:T547K, S:N856K, and S:L981F, and BA.2 contains S:V213G, S:T376A, and S:D405N. Notably, ins214EPE (S1- N-Terminal domain) mutation was found in a significant number of Omicron BA.1 and sub-lineages. The overall Omicron BA.2 lineage was observed in 79.5% of women and 83.2% of men.
Conclusion: The current study showed a predominance of the Omicron BA.2 variant outcompeting the BA.1 over a period in eastern UP. Most of the cases had a breakthrough infection following the recommended two doses of vaccine with four in five cases being symptomatic. There is a need to further explore the immune evasion properties of the Omicron variant.
Keywords: COVID-19; SARS-CoV-2; omicron; third wave; variant of concern (VOC); whole-genome sequencing (WGS).
Copyright © 2022 Zaman, Shete, Mishra, Kumar, Reddy, Sahay, Yadav, Majumdar, Pandey, Dwivedi, Deval, Singh, Behera, Kumar, Patil, Kumar, Dudhmal, Joshi, Shukla, Gawande, Kavathekar, Kumar, Kumar, Kumar, Singh, Kumar, Tiwari, Verma, Yadav and Kant.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- WHO. Coronavirus (COVID-19) Dashboard. Geneva: World Health Organization; (2022).
-
- MoHFW. COVID-19 Statewise Status. New Delhi: Ministry of Health and Family Welfare, Government of India; (2022).
LinkOut - more resources
Full Text Sources
Miscellaneous

