Single cell analysis of docosahexaenoic acid suppression of sequential LPS-induced proinflammatory and interferon-regulated gene expression in the macrophage
- PMID: 36405730
- PMCID: PMC9669445
- DOI: 10.3389/fimmu.2022.993614
Single cell analysis of docosahexaenoic acid suppression of sequential LPS-induced proinflammatory and interferon-regulated gene expression in the macrophage
Abstract
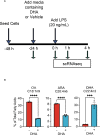
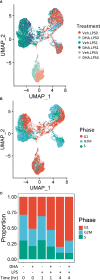
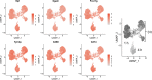
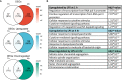
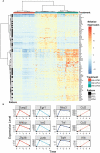
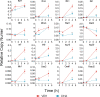
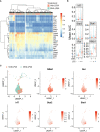
Preclinical and clinical studies suggest that consumption of long chain omega-3 polyunsaturated fatty acids (PUFAs) reduces severity of chronic inflammatory and autoimmune diseases. While these ameliorative effects are conventionally associated with downregulated expression of proinflammatory cytokine and chemokine genes, our laboratory has recently identified Type 1 interferon (IFN1)-regulated gene expression to be another key target of omega-3 PUFAs. Here we used single cell RNA sequencing (scRNAseq) to gain new mechanistic perspectives on how the omega-3 PUFA docosahexaenoic acid (DHA) influences TLR4-driven proinflammatory and IFN1-regulated gene expression in a novel self-renewing murine fetal liver-derived macrophage (FLM) model. FLMs were cultured with 25 µM DHA or vehicle for 24 h, treated with modest concentration of LPS (20 ng/ml) for 1 and 4 h, and then subjected to scRNAseq using the 10X Chromium System. At 0 h (i.e., in the absence of LPS), DHA increased expression of genes associated with the NRF2 antioxidant response (e.g. Sqstm1, Hmox1, Chchd10) and metal homeostasis (e.g.Mt1, Mt2, Ftl1, Fth1), both of which are consistent with DHA-induced polarization of FLMs to a more anti-inflammatory phenotype. At 1 h post-LPS treatment, DHA inhibited LPS-induced cholesterol synthesis genes (e.g. Scd1, Scd2, Pmvk, Cyp51, Hmgcs1, and Fdps) which potentially could contribute to interference with TLR4-mediated inflammatory signaling. At 4 h post-LPS treatment, LPS-treated FLMs reflected a more robust inflammatory response including upregulation of proinflammatory cytokine (e.g. Il1a, Il1b, Tnf) and chemokine (e.g.Ccl2, Ccl3, Ccl4, Ccl7) genes as well as IFN1-regulated genes (e.g. Irf7, Mx1, Oasl1, Ifit1), many of which were suppressed by DHA. Using single-cell regulatory network inference and clustering (SCENIC) to identify gene expression networks, we found DHA modestly downregulated LPS-induced expression of NF-κB-target genes. Importantly, LPS induced a subset of FLMs simultaneously expressing NF-κB- and IRF7/STAT1/STAT2-target genes that were conspicuously absent in DHA-pretreated FLMs. Thus, DHA potently targeted both the NF-κB and the IFN1 responses. Altogether, scRNAseq generated a valuable dataset that provides new insights into multiple overlapping mechanisms by which DHA may transcriptionally or post-transcriptionally regulate LPS-induced proinflammatory and IFN1-driven responses in macrophages.
Keywords: IFN signaling; NF-κB; TLR4 signaling; cholesterol metabolism; inflammatory gene expression; macrophage; omega-3 polyunsaturated fatty acids; scRNAseq.
Copyright © 2022 Wierenga, Riemers, Westendorp, Harkema and Pestka.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Crystalline silica-induced proinflammatory eicosanoid storm in novel alveolar macrophage model quelled by docosahexaenoic acid supplementation.Front Immunol. 2023 Nov 9;14:1274147. doi: 10.3389/fimmu.2023.1274147. eCollection 2023. Front Immunol. 2023. PMID: 38022527 Free PMC article.
-
Docosahexaenoic acid induces an anti-inflammatory profile in lipopolysaccharide-stimulated human THP-1 macrophages more effectively than eicosapentaenoic acid.J Nutr Biochem. 2007 Apr;18(4):250-8. doi: 10.1016/j.jnutbio.2006.04.003. Epub 2006 Jun 16. J Nutr Biochem. 2007. PMID: 16781858
-
EPA and DHA reduce LPS-induced inflammation responses in HK-2 cells: evidence for a PPAR-gamma-dependent mechanism.Kidney Int. 2005 Mar;67(3):867-74. doi: 10.1111/j.1523-1755.2005.00151.x. Kidney Int. 2005. PMID: 15698426
-
Docosahexaenoic acid attenuates LPS-stimulated inflammatory response by regulating the PPARγ/NF-κB pathways in primary bovine mammary epithelial cells.Res Vet Sci. 2017 Jun;112:7-12. doi: 10.1016/j.rvsc.2016.12.011. Epub 2017 Jan 5. Res Vet Sci. 2017. PMID: 28095338 Review.
-
Immunomodulatory Effects of Omega-3 Fatty Acids: Mechanistic Insights and Health Implications.Mol Nutr Food Res. 2025 May;69(10):e202400752. doi: 10.1002/mnfr.202400752. Epub 2025 Mar 30. Mol Nutr Food Res. 2025. PMID: 40159804 Free PMC article. Review.
Cited by
-
Dietary docosahexaenoic acid supplementation inhibits acute pulmonary transcriptional and autoantibody responses to a single crystalline silica exposure in lupus-prone mice.Front Immunol. 2024 Feb 1;15:1275265. doi: 10.3389/fimmu.2024.1275265. eCollection 2024. Front Immunol. 2024. PMID: 38361937 Free PMC article.
-
The Impact of Polyunsaturated Fatty Acids in Cancer and Therapeutic Strategies.Curr Nutr Rep. 2025 Mar 14;14(1):46. doi: 10.1007/s13668-025-00639-y. Curr Nutr Rep. 2025. PMID: 40085324 Review.
-
Regulation of macrophage polarization by targeted metabolic reprogramming for the treatment of lupus nephritis.Mol Med. 2024 Jun 25;30(1):96. doi: 10.1186/s10020-024-00866-z. Mol Med. 2024. PMID: 38914953 Free PMC article. Review.
-
Crystalline silica-induced proinflammatory eicosanoid storm in novel alveolar macrophage model quelled by docosahexaenoic acid supplementation.Front Immunol. 2023 Nov 9;14:1274147. doi: 10.3389/fimmu.2023.1274147. eCollection 2023. Front Immunol. 2023. PMID: 38022527 Free PMC article.
-
Lipidome modulation by dietary omega-3 polyunsaturated fatty acid supplementation or selective soluble epoxide hydrolase inhibition suppresses rough LPS-accelerated glomerulonephritis in lupus-prone mice.Front Immunol. 2023 Feb 16;14:1124910. doi: 10.3389/fimmu.2023.1124910. eCollection 2023. Front Immunol. 2023. PMID: 36875087 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

