Identifying hub genes of calcific aortic valve disease and revealing the immune infiltration landscape based on multiple WGCNA and single-cell sequence analysis
- PMID: 36405745
- PMCID: PMC9673246
- DOI: 10.3389/fimmu.2022.1035285
Identifying hub genes of calcific aortic valve disease and revealing the immune infiltration landscape based on multiple WGCNA and single-cell sequence analysis
Abstract
Background: Calcific aortic valve disease (CAVD) is a progressive fibrocalcific disease that can be treated only through valve replacement. This study aimed to determine the role of hub genes and immune cell infiltration in CAVD progression.
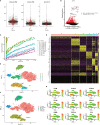
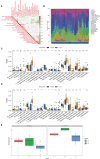
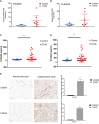
Methods: In this study, bioinformatics analysis was used to identify hub genes involved in CAVD. The datasets were downloaded from the Gene Expression Omnibus (GEO) database. Gene expression differences were evaluated via pathway and Gene Ontology analyses. Weighted gene co-expression network analysis (WGCNA) and differentially expressed genes were used to screen hub genes. The CIBERSORT algorithm was used to compare immune infiltration into the calcified aortic valve based on the hub genes between high- and low-expression groups. We also performed single-cell RNA sequencing based on six different human aortic valve leaflets. The expression of hub genes was identified in human and mouse samples through quantitative real-time polymerase chain reaction (qPCR), immunohistochemistry, immunofluorescence, and ELISA, and clinical features of the patients were investigated.
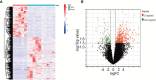
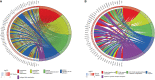
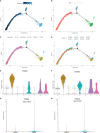
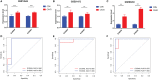
Results: In total, 454 differentially expressed genes were obtained from the GEO database. WGCNA was used to find 12 co-expression modules in the Array Express database, of which one hub module (brown module) was most correlated with CAVD. Two hub genes were identified after combining the differentially expressed genes S100A8 and S100A9. Regarding these genes, the immune infiltration profiles varied between high- and low-expression groups. Compared with that in the low hub gene expression group, the high hub gene expression group had a higher proportion of activated NK cells (p < 0.01) and M1 macrophages (p < 0.05). The expression of S100A8 and S100A9 was consistent with single-gene RNA sequencing results, confirming that the expression levels of these two hub genes are significantly upregulated in patients with CAVD (p < 0.01). Furthermore, these results were verified using mouse and human samples by performing immunofluorescence, immunohistochemistry, qPCR, and ELISA analyses. Finally, the localization of S100A8 and S100A9 in monocytes and macrophages was confirmed via immunofluorescence using human aortic valves.
Conclusion: These results demonstrate that S100A8 and S100A9 are two hub genes involved in CAVD, which might play an important role in its development through immune-related signaling pathways.
Keywords: DEGs; bioinformatics; calcific aortic valve diseases (CAVD); immune infiltration; single cell sequence (scRNA-seq); weighted gene co-expression network analysis (WGCNA).
Copyright © 2022 Wang, Zheng, Liu, Geng, Dong and Shi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

