Targeted proteomics identifies circulating biomarkers associated with active COVID-19 and post-COVID-19
- PMID: 36405747
- PMCID: PMC9670186
- DOI: 10.3389/fimmu.2022.1027122
Targeted proteomics identifies circulating biomarkers associated with active COVID-19 and post-COVID-19
Abstract
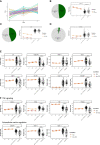
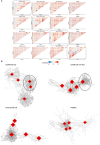
The ongoing Coronavirus Disease 2019 (COVID-19) pandemic is caused by the highly infectious Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2). There is an urgent need for biomarkers that will help in better stratification of patients and contribute to personalized treatments. We performed targeted proteomics using the Olink platform and systematically investigated protein concentrations in 350 hospitalized COVID-19 patients, 186 post-COVID-19 individuals, and 61 healthy individuals from 3 independent cohorts. Results revealed a signature of acute SARS-CoV-2 infection, which is represented by inflammatory biomarkers, chemokines and complement-related factors. Furthermore, the circulating proteome is still significantly affected in post-COVID-19 samples several weeks after infection. Post-COVID-19 individuals are characterized by upregulation of mediators of the tumor necrosis (TNF)-α signaling pathways and proteins related to transforming growth factor (TGF)-ß. In addition, the circulating proteome is able to differentiate between patients with different COVID-19 disease severities, and is associated with the time after infection. These results provide important insights into changes induced by SARS-CoV-2 infection at the proteomic level by integrating several cohorts to obtain a large disease spectrum, including variation in disease severity and time after infection. These findings could guide the development of host-directed therapy in COVID-19.
Keywords: SARS-CoV-2; biomarker; inflammation; post-COVID-19; targeted proteomics.
Copyright © 2022 Zoodsma, de Nooijer, Grondman, Gupta, Bonifacius, Koeken, Kooistra, Kilic, Bulut, Gödecke, Janssen, Kox, Domínguez-Andrés, van Gammeren, Ermens, van der Ven, Pickkers, Blasczyk, Behrens, van de Veerdonk, Joosten, Xu, Eiz-Vesper, Netea and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Plasma proteomic signature predicts who will get persistent symptoms following SARS-CoV-2 infection.EBioMedicine. 2022 Nov;85:104293. doi: 10.1016/j.ebiom.2022.104293. Epub 2022 Sep 28. EBioMedicine. 2022. PMID: 36182629 Free PMC article.
-
Plasma proteomics identify potential severity biomarkers from COVID-19 associated network.Proteomics Clin Appl. 2023 Mar;17(2):e2200070. doi: 10.1002/prca.202200070. Epub 2022 Oct 25. Proteomics Clin Appl. 2023. PMID: 36217943 Free PMC article.
-
Virus-Host Interactome and Proteomic Survey Reveal Potential Virulence Factors Influencing SARS-CoV-2 Pathogenesis.Med. 2021 Jan 15;2(1):99-112.e7. doi: 10.1016/j.medj.2020.07.002. Epub 2020 Jul 21. Med. 2021. PMID: 32838362 Free PMC article.
-
Progress in understanding COVID-19: insights from the omics approach.Crit Rev Clin Lab Sci. 2021 Jun;58(4):242-252. doi: 10.1080/10408363.2020.1851167. Epub 2020 Dec 29. Crit Rev Clin Lab Sci. 2021. PMID: 33375876 Review.
-
Comprehensive proteomics and meta-analysis of COVID-19 host response.Nat Commun. 2023 Sep 22;14(1):5921. doi: 10.1038/s41467-023-41159-z. Nat Commun. 2023. PMID: 37739942 Free PMC article.
Cited by
-
Distinct proteomic signatures in Ethiopians predict acute and long-term sequelae of COVID-19.Front Immunol. 2025 May 22;16:1575135. doi: 10.3389/fimmu.2025.1575135. eCollection 2025. Front Immunol. 2025. PMID: 40475767 Free PMC article.
-
COVID-19-Omics Report: From Individual Omics Approaches to Precision Medicine.Reports (MDPI). 2023 Sep 22;6(4):45. doi: 10.3390/reports6040045. Reports (MDPI). 2023. PMID: 40729168 Free PMC article. Review.
-
Proteomic profiling identifies biomarkers of COVID-19 severity.Heliyon. 2023 Dec 9;10(1):e23320. doi: 10.1016/j.heliyon.2023.e23320. eCollection 2024 Jan 15. Heliyon. 2023. PMID: 38163173 Free PMC article.
-
Identification of shared biological features in four different lung cell lines infected with SARS-CoV-2 virus through RNA-seq analysis.Front Genet. 2023 Aug 16;14:1235927. doi: 10.3389/fgene.2023.1235927. eCollection 2023. Front Genet. 2023. PMID: 37662846 Free PMC article.
-
A Review on Long COVID Screening: Challenges and Perspectives Focusing on Exhaled Breath Gas Sensing.ACS Sens. 2025 Mar 28;10(3):1564-1578. doi: 10.1021/acssensors.4c02280. Epub 2024 Dec 16. ACS Sens. 2025. PMID: 39680873 Free PMC article. Review.
References
-
- World Health Organization . (2020)., in: Virtual press conference on COVID-19, , 11 March 2020. Available at: https://www.who.int/docs/default-source/coronaviruse/transcripts/who-aud....
-
- Ahmed H, Patel K, Greenwood DC, Halpin S, Lewthwaite P, Salawu A, et al. . Long-term clinical outcomes in survivors of severe acute respiratory syndrome (SARS) and middle East respiratory syndrome coronavirus (MERS) outbreaks after hospitalisation or ICU admission: a systematic review and meta-analysis. J Rehabil Med (2020) 52(5):1–11. doi: 10.2340/16501977-2694 - DOI - PubMed
-
- Guler SA, Ebner L, Beigelman C, Bridevaux PO, Brutsche M, Clarenbach C, et al. . Pulmonary function and radiological features four months after COVID-19: first results from the national prospective observational Swiss COVID-19 lung study. Eur Respir J (2021) 57:2003690. doi: 10.1183/13993003.03690-2020 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

