Establishment and analysis of a disease risk prediction model for the systemic lupus erythematosus with random forest
- PMID: 36405750
- PMCID: PMC9667742
- DOI: 10.3389/fimmu.2022.1025688
Establishment and analysis of a disease risk prediction model for the systemic lupus erythematosus with random forest
Abstract
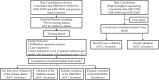
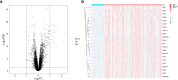
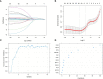
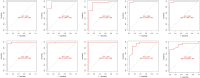
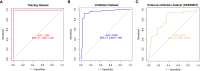
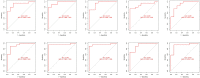
Systemic lupus erythematosus (SLE) is a latent, insidious autoimmune disease, and with the development of gene sequencing in recent years, our study aims to develop a gene-based predictive model to explore the identification of SLE at the genetic level. First, gene expression datasets of SLE whole blood samples were collected from the Gene Expression Omnibus (GEO) database. After the datasets were merged, they were divided into training and validation datasets in the ratio of 7:3, where the SLE samples and healthy samples of the training dataset were 334 and 71, respectively, and the SLE samples and healthy samples of the validation dataset were 143 and 30, respectively. The training dataset was used to build the disease risk prediction model, and the validation dataset was used to verify the model identification ability. We first analyzed differentially expressed genes (DEGs) and then used Lasso and random forest (RF) to screen out six key genes (OAS3, USP18, RTP4, SPATS2L, IFI27 and OAS1), which are essential to distinguish SLE from healthy samples. With six key genes incorporated and five iterations of 10-fold cross-validation performed into the RF model, we finally determined the RF model with optimal mtry. The mean values of area under the curve (AUC) and accuracy of the models were over 0.95. The validation dataset was then used to evaluate the AUC performance and our model had an AUC of 0.948. An external validation dataset (GSE99967) with an AUC of 0.810, an accuracy of 0.836, and a sensitivity of 0.921 was used to assess the model's performance. The external validation dataset (GSE185047) of all SLE patients yielded an SLE sensitivity of up to 0.954. The final high-throughput RF model had a mean value of AUC over 0.9, again showing good results. In conclusion, we identified key genetic biomarkers and successfully developed a novel disease risk prediction model for SLE that can be used as a new SLE disease risk prediction aid and contribute to the identification of SLE.
Keywords: GEO; Lasso; disease risk prediction model; random forest; systemic lupus erythematosus.
Copyright © 2022 Chen, Huang, Jiang, Wang, Bian, Ma and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Decoding the mitochondrial connection: development and validation of biomarkers for classifying and treating systemic lupus erythematosus through bioinformatics and machine learning.BMC Rheumatol. 2023 Dec 4;7(1):44. doi: 10.1186/s41927-023-00369-0. BMC Rheumatol. 2023. PMID: 38044432 Free PMC article.
-
Development and evaluation of a chronic kidney disease risk prediction model using random forest.Front Genet. 2024 Jun 27;15:1409755. doi: 10.3389/fgene.2024.1409755. eCollection 2024. Front Genet. 2024. PMID: 38993480 Free PMC article.
-
Establishment and analysis of a novel diagnostic model for systemic juvenile idiopathic arthritis based on machine learning.Pediatr Rheumatol Online J. 2024 Jan 19;22(1):18. doi: 10.1186/s12969-023-00949-x. Pediatr Rheumatol Online J. 2024. PMID: 38243323 Free PMC article.
-
Development and validation of a risk calculator to differentiate flares from infections in systemic lupus erythematosus patients with fever.Autoimmun Rev. 2015 Jul;14(7):586-93. doi: 10.1016/j.autrev.2015.02.005. Epub 2015 Feb 20. Autoimmun Rev. 2015. PMID: 25703012 Review.
-
From incomplete to complete systemic lupus erythematosus; A review of the predictive serological immune markers.Semin Arthritis Rheum. 2021 Feb;51(1):43-48. doi: 10.1016/j.semarthrit.2020.11.006. Epub 2020 Dec 18. Semin Arthritis Rheum. 2021. PMID: 33360229 Review.
Cited by
-
Decoding the mitochondrial connection: development and validation of biomarkers for classifying and treating systemic lupus erythematosus through bioinformatics and machine learning.BMC Rheumatol. 2023 Dec 4;7(1):44. doi: 10.1186/s41927-023-00369-0. BMC Rheumatol. 2023. PMID: 38044432 Free PMC article.
-
SPATS2L is a positive feedback regulator of the type I interferon signaling pathway and plays a vital role in lupus.Acta Biochim Biophys Sin (Shanghai). 2024 Aug 2;56(11):1659-1672. doi: 10.3724/abbs.2024132. Acta Biochim Biophys Sin (Shanghai). 2024. PMID: 39099414 Free PMC article.
-
Multi-omics analysis reveals interferon-stimulated gene OAS1 as a prognostic and immunological biomarker in pan-cancer.Front Immunol. 2023 Oct 20;14:1249731. doi: 10.3389/fimmu.2023.1249731. eCollection 2023. Front Immunol. 2023. PMID: 37928544 Free PMC article.
-
Development and evaluation of a chronic kidney disease risk prediction model using random forest.Front Genet. 2024 Jun 27;15:1409755. doi: 10.3389/fgene.2024.1409755. eCollection 2024. Front Genet. 2024. PMID: 38993480 Free PMC article.
-
Use of Machine Learning for the Identification and Validation of Immunogenic Cell Death Biomarkers and Immunophenotypes in Coronary Artery Disease.J Inflamm Res. 2024 Jan 12;17:223-249. doi: 10.2147/JIR.S439315. eCollection 2024. J Inflamm Res. 2024. PMID: 38229693 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

