Why not try to predict autism spectrum disorder with crucial biomarkers in cuproptosis signaling pathway?
- PMID: 36405901
- PMCID: PMC9667021
- DOI: 10.3389/fpsyt.2022.1037503
Why not try to predict autism spectrum disorder with crucial biomarkers in cuproptosis signaling pathway?
Abstract
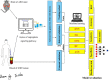
The exact pathogenesis of autism spectrum disorder (ASD) is still unclear, yet some potential mechanisms may not have been evaluated before. Cuproptosis is a novel form of regulated cell death reported this year, and no study has reported the relationship between ASD and cuproptosis. This study aimed to identify ASD in suspected patients early using machine learning models based on biomarkers of the cuproptosis pathway. We collected gene expression profiles from brain samples from ASD model mice and blood samples from humans with ASD, selected crucial genes in the cuproptosis signaling pathway, and then analysed these genes with different machine learning models. The accuracy, sensitivity, specificity, and areas under the receiver operating characteristic curves of the machine learning models were estimated in the training, internal validation, and external validation cohorts. Differences between models were determined with Bonferroni's test. The results of screening with the Boruta algorithm showed that FDX1, DLAT, LIAS, and ATP7B were crucial genes in the cuproptosis signaling pathway for ASD. All selected genes and corresponding proteins were also expressed in the human brain. The k-nearest neighbor, support vector machine and random forest models could identify approximately 72% of patients with ASD. The artificial neural network (ANN) model was the most suitable for the present data because the accuracy, sensitivity, and specificity were 0.90, 1.00, and 0.80, respectively, in the external validation cohort. Thus, we first report the prediction of ASD in suspected patients with machine learning methods based on crucial biomarkers in the cuproptosis signaling pathway, and these findings may contribute to investigations of the potential pathogenesis and early identification of ASD.
Keywords: artificial neural network; autism spectrum disorder; biomarkers; cuproptosis; machine learning.
Copyright © 2022 Zhou and Gao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Machine learning-based identification of cuproptosis-related markers and immune infiltration in severe community-acquired pneumonia.Clin Respir J. 2023 Jul;17(7):618-628. doi: 10.1111/crj.13633. Epub 2023 Jun 6. Clin Respir J. 2023. PMID: 37279744 Free PMC article.
-
Identification and immunological characterization of cuproptosis-related molecular clusters in Alzheimer's disease.Front Aging Neurosci. 2022 Jul 28;14:932676. doi: 10.3389/fnagi.2022.932676. eCollection 2022. Front Aging Neurosci. 2022. PMID: 35966780 Free PMC article.
-
Regulation, genomics, and clinical characteristics of cuproptosis regulators in pan-cancer.Front Oncol. 2022 Oct 27;12:934076. doi: 10.3389/fonc.2022.934076. eCollection 2022. Front Oncol. 2022. PMID: 36387247 Free PMC article.
-
The implications and prospect of cuproptosis-related genes and copper transporters in cancer progression.Front Oncol. 2023 Feb 28;13:1117164. doi: 10.3389/fonc.2023.1117164. eCollection 2023. Front Oncol. 2023. PMID: 36925927 Free PMC article. Review.
-
Food for Thought: Machine Learning in Autism Spectrum Disorder Screening of Infants.Cureus. 2021 Oct 12;13(10):e18721. doi: 10.7759/cureus.18721. eCollection 2021 Oct. Cureus. 2021. PMID: 34790476 Free PMC article. Review.
Cited by
-
Mitochondrial pathways of copper neurotoxicity: focus on mitochondrial dynamics and mitophagy.Front Mol Neurosci. 2024 Dec 5;17:1504802. doi: 10.3389/fnmol.2024.1504802. eCollection 2024. Front Mol Neurosci. 2024. PMID: 39703721 Free PMC article. Review.
-
Identification of Immune Infiltration and Iron Metabolism-Related Subgroups in Autism Spectrum Disorder.J Mol Neurosci. 2024 Jan 18;74(1):12. doi: 10.1007/s12031-023-02179-y. J Mol Neurosci. 2024. PMID: 38236354
References
LinkOut - more resources
Full Text Sources

