The nitric oxide synthase gene negatively regulates biofilm formation in Staphylococcus epidermidis
- PMID: 36405963
- PMCID: PMC9669438
- DOI: 10.3389/fcimb.2022.1015859
The nitric oxide synthase gene negatively regulates biofilm formation in Staphylococcus epidermidis
Abstract
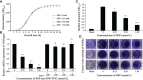
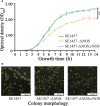
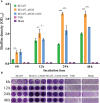
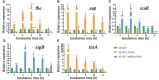
Staphylococcus epidermidis (S. epidermidis) is a clinically important conditioned pathogen that can cause a troublesome chronic implant-related infection once a biofilm is formed. The nitric oxide synthase (NOS) gene, which is responsible for endogenous nitric oxide synthesis, has already been found in the genome of S. epidermidis; however, the specific mechanisms associated with the effects of NOS on S. epidermidis pathogenicity are still unknown. The purpose of the current study was to investigate whether the NOS gene has an impact on biofilm formation in S. epidermidis. Bioinformatics analysis of the NOS gene was performed, and homologous recombination was subsequently employed to delete this gene. The effects of the NOS gene on biofilm formation of S. epidermidis and its underlying mechanisms were analyzed by bacterial growth assays, biofilm semiquantitative determination, Triton X-100-induced autolysis assays, and bacterial biofilm dispersal assays. Additionally, the transcription levels of fbe, aap, icaA, icaR and sigB, which are related to biofilm formation, were further investigated by qRT-PCR following NOS deletion. Phylogenetic analysis revealed that the NOS gene was conserved between bacterial species originating from different genera. The NOS deletion strain of S. epidermidis 1457 and its counterpart were successfully constructed. Disruption of the NOS gene resulted in significantly enhanced biofilm formation, slightly retarded bacterial growth, a markedly decreased autolysis rate, and drastically weakened bacterial biofilm dispersal. Our data showed that the fbe, aap and icaA genes were significantly upregulated, while the icaR and sigB genes were significantly downregulated, compared with the wild strain. Therefore, these data strongly suggested that the NOS gene can negatively regulate biofilm formation in S. epidermidis by affecting biofilm aggregation and dispersal.
Keywords: S. epidermidis; biofilm; chronic infection; nitric oxide; nitric oxide synthase gene.
Copyright © 2022 Wang, Rao, Huang, Ma, Yang, Yu, Sun and Ge.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer FY declared a past co-authorship with the author LR to the handling editor.
Figures


Similar articles
-
The Vancomycin Resistance-Associated Regulatory System VraSR Modulates Biofilm Formation of Staphylococcus epidermidis in an ica-Dependent Manner.mSphere. 2021 Oct 27;6(5):e0064121. doi: 10.1128/mSphere.00641-21. Epub 2021 Sep 22. mSphere. 2021. PMID: 34550006 Free PMC article.
-
Mechanistic study on the inhibition of Staphylococcus epidermidis biofilm by agrC-specific binding polypeptide.Ann Transl Med. 2020 Mar;8(6):337. doi: 10.21037/atm.2020.02.84. Ann Transl Med. 2020. PMID: 32355781 Free PMC article.
-
Role of the SaeRS two-component regulatory system in Staphylococcus epidermidis autolysis and biofilm formation.BMC Microbiol. 2011 Jun 24;11:146. doi: 10.1186/1471-2180-11-146. BMC Microbiol. 2011. PMID: 21702925 Free PMC article.
-
Polysaccharide intercellular adhesin in biofilm: structural and regulatory aspects.Front Cell Infect Microbiol. 2015 Feb 10;5:7. doi: 10.3389/fcimb.2015.00007. eCollection 2015. Front Cell Infect Microbiol. 2015. PMID: 25713785 Free PMC article. Review.
-
Structural basis of Staphylococcus epidermidis biofilm formation: mechanisms and molecular interactions.Front Cell Infect Microbiol. 2015 Feb 17;5:14. doi: 10.3389/fcimb.2015.00014. eCollection 2015. Front Cell Infect Microbiol. 2015. PMID: 25741476 Free PMC article. Review.
Cited by
-
The first genomic characterization of a stable, hemin-dependent small colony variant strain of Staphylococcus epidermidis isolated from a prosthetic-joint infection.Front Microbiol. 2023 Oct 19;14:1289844. doi: 10.3389/fmicb.2023.1289844. eCollection 2023. Front Microbiol. 2023. PMID: 37928677 Free PMC article.
References
-
- Christner M., Heinze C., Busch M., Franke G., Hentschke M., Bayard Dühring S., et al. . (2012). sarA negatively regulates Staphylococcus epidermidis biofilm formation by modulating expression of 1 MDa extracellular matrix binding protein and autolysis-dependent release of eDNA. Mol. Microbiol. 86 (2), 394–410. doi: 10.1111/j.1365-2958.2012.08203.x - DOI - PubMed
-
- Conlon B. P., Geoghegan J. A., Waters E. M., McCarthy H., Rowe S. E., Davies J. R., et al. . (2014). Role for the a domain of unprocessed accumulation-associated protein (Aap) in the attachment phase of the Staphylococcus epidermidis biofilm phenotype. J. Bacteriol. 196 (24), 4268–4275. doi: 10.1128/JB.01946-14 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

