MicroRNA and circular RNA profiling in the deposited fat tissue of Sunite sheep
- PMID: 36406061
- PMCID: PMC9672515
- DOI: 10.3389/fvets.2022.954882
MicroRNA and circular RNA profiling in the deposited fat tissue of Sunite sheep
Abstract
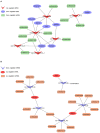
As the most typical deposited fat, tail fat is an important energy reservoir for sheep adapted to harsh environments and plays an important role as a raw material in daily life. However, the regulatory mechanisms of microRNA (miRNA) and circular RNA (circRNA) in tail fat development remain unclear. In this study, we characterized the miRNA and circRNA expression profiles in the tail fat of sheep at the ages of 6, 18, and 30 months. We identified 219 differentially expressed (DE) miRNAs (including 12 novel miRNAs), which exhibited a major tendency to be downregulated, and 198 DE circRNAs, which exhibited a tendency to be upregulated. Target gene prediction analysis was performed for the DE miRNAs. Functional analysis revealed that their target genes were mainly involved in cellular interactions, while the host genes of DE circRNAs were implicated in lipid and fatty acid metabolism. Subsequently, we established a competing endogenous RNA (ceRNA) network based on the negative regulatory relationship between miRNAs and target genes. The network revealed that upregulated miRNAs play a leading role in the development of tail fat. Finally, the ceRNA relationship network with oar-miR-27a_R-1 and oar-miR-29a as the core was validated, suggesting possible involvement of these interactions in tail fat development. In summary, DE miRNAs were negatively correlated with DE circRNAs during sheep tail fat development. The multiple ceRNA regulatory network dominated by upregulated DE miRNAs may play a key role in this developmental process.
Keywords: Sunite sheep; circular RNA; competing endogenous RNA; microRNA; tail fat.
Copyright © 2022 He, Wu, Yun, Qin, Huang, Chen, Han, Wu, Sha and Borjigin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Identification of Serum Exosome-Derived circRNA-miRNA-TF-mRNA Regulatory Network in Postmenopausal Osteoporosis Using Bioinformatics Analysis and Validation in Peripheral Blood-Derived Mononuclear Cells.Front Endocrinol (Lausanne). 2022 Jun 9;13:899503. doi: 10.3389/fendo.2022.899503. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35757392 Free PMC article.
-
Functional analysis of differentially expressed circular RNAs in sheep subcutaneous fat.BMC Genomics. 2023 Oct 5;24(1):591. doi: 10.1186/s12864-023-09401-6. BMC Genomics. 2023. PMID: 37798722 Free PMC article.
-
Transcriptome analysis of messenger RNA and long noncoding RNA related to different developmental stages of tail adipose tissues of sunite sheep.Food Sci Nutr. 2021 Aug 25;9(10):5722-5734. doi: 10.1002/fsn3.2537. eCollection 2021 Oct. Food Sci Nutr. 2021. PMID: 34646540 Free PMC article.
-
RNA-Seq Revealed a Circular RNA-microRNA-mRNA Regulatory Network in Hantaan Virus Infection.Front Cell Infect Microbiol. 2020 Mar 13;10:97. doi: 10.3389/fcimb.2020.00097. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32232013 Free PMC article. Review.
-
Deciphering the role of alternative splicing as a potential regulator in fat-tail development of sheep: a comprehensive RNA-seq based study.Sci Rep. 2024 Jan 29;14(1):2361. doi: 10.1038/s41598-024-52855-1. Sci Rep. 2024. PMID: 38287039 Free PMC article. Review.
Cited by
-
Mechanisms of Adipose Tissue Metabolism in Naturally Grazing Sheep at Different Growth Stages: Insights from mRNA and miRNA Profiles.Int J Mol Sci. 2025 Apr 2;26(7):3324. doi: 10.3390/ijms26073324. Int J Mol Sci. 2025. PMID: 40244192 Free PMC article.
-
The Characterization of Subcutaneous Adipose Tissue in Sunit Sheep at Different Growth Stages: A Comprehensive Analysis of the Morphology, Fatty Acid Profile, and Metabolite Profile.Foods. 2024 Feb 9;13(4):544. doi: 10.3390/foods13040544. Foods. 2024. PMID: 38397521 Free PMC article.
-
Understanding Circular RNAs in Health, Welfare, and Productive Traits of Cattle, Goats, and Sheep.Animals (Basel). 2024 Feb 27;14(5):733. doi: 10.3390/ani14050733. Animals (Basel). 2024. PMID: 38473119 Free PMC article. Review.
-
Profile of miRNAs induced during sheep fat tail development and roles of four key miRNAs in proliferation and differentiation of sheep preadipocytes.Front Vet Sci. 2024 Dec 3;11:1491160. doi: 10.3389/fvets.2024.1491160. eCollection 2024. Front Vet Sci. 2024. PMID: 39691379 Free PMC article.
-
Quantitative Proteomics Revealed the Molecular Regulatory Network of Lysine and the Effects of Lysine Supplementation on Sunit Skeletal Muscle Satellite Cells.Animals (Basel). 2025 May 14;15(10):1425. doi: 10.3390/ani15101425. Animals (Basel). 2025. PMID: 40427302 Free PMC article.
References
-
- Kashan NE J, Azar GHM, Afzalzadeh A, Salehi A. Growth performance and carcass quality of fattening lambs from fat-tailed and tailed sheep breeds. Small Ruminant Res. (2005) 60:267–71. 10.1016/j.smallrumres.2005.01.001 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

