Non-invasive prenatal testing for autosomal recessive disorders: A new promising approach
- PMID: 36406136
- PMCID: PMC9669374
- DOI: 10.3389/fgene.2022.1047474
Non-invasive prenatal testing for autosomal recessive disorders: A new promising approach
Abstract
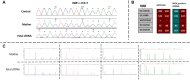
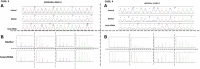
Background: In pregnant women at risk of autosomal recessive (AR) disorders, prenatal diagnosis of AR disorders primarily involves invasive procedures, such as chorionic villus sampling and amniocentesis. Methods: We collected blood samples from four pregnant women in their first trimester who presented a risk of having a child with an AR disorder. Cell-free DNA (cfDNA) was extracted, amplified, and double-purified to reduce maternal DNA interference. Additionally, whole-genome amplification was performed for traces of residual purified cfDNA for utilization in subsequent applications. Results: Based on our findings, we detected the fetal status with the family corresponding different genes, i.e., LZTR1, DVL2, HBB, RNASEH2B, and MYO7A, as homozygous affected, wild-type, and heterozygous carriers, respectively. Results were subsequently confirmed by prenatal amniocentesis. The results of AmpFLSTR™ Identifiler™ presented a distinct profile from the corresponding mother profile, thereby corroborating the result reflecting the genetic material of the fetus. Conclusion: Herein, we detected AR disease mutations in the first trimester of pregnancy while surmounting limitations associated with maternal genetic material interference. Importantly, such detection strategies would allow the screening of pregnant women for common AR diseases, especially in highly consanguineous marriage populations. This technique would open avenues for the early detection and prevention of recessive diseases among the population.
Keywords: AR; Single gene; autosomal recessive; fetal DNA; non-invasive prenatal testing for monogenic disorders (NIPTM); whole-genome amplification.
Copyright © 2022 Alyafee, Al Tuwaijri, Umair, Alharbi, Haddad, Ballow, Alayyar, Alam, Althenayyan, Al Ghilan, Al Khaldi, Faden, Al Sufyan and Alfadhel.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Research Materials

