Multifaceted Influence of Histone Deacetylases on DNA Damage Repair: Implications for Hepatocellular Carcinoma
- PMID: 36406320
- PMCID: PMC9647118
- DOI: 10.14218/JCTH.2022.00079
Multifaceted Influence of Histone Deacetylases on DNA Damage Repair: Implications for Hepatocellular Carcinoma
Abstract
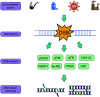
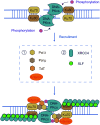
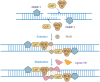
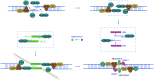
Hepatocellular carcinoma (HCC) is one of the most commonly diagnosed cancers and a leading cause of cancer-related mortality worldwide, but its pathogenesis remains largely unknown. Nevertheless, genomic instability has been recognized as one of the facilitating characteristics of cancer hallmarks that expedites the acquisition of genetic diversity. Genomic instability is associated with a greater tendency to accumulate DNA damage and tumor-specific DNA repair defects, which gives rise to gene mutations and chromosomal damage and causes oncogenic transformation and tumor progression. Histone deacetylases (HDACs) have been shown to impair a variety of cellular processes of genome stability, including the regulation of DNA damage and repair, reactive oxygen species generation and elimination, and progression to mitosis. In this review, we provide an overview of the role of HDAC in the different aspects of DNA repair and genome instability in HCC as well as the current progress on the development of HDAC-specific inhibitors as new cancer therapies.
Keywords: DNA repair; Hepatocellular carcinoma; Histone deacetylases.
© 2023 Authors.
Conflict of interest statement
The authors have no conflict of interests related to this publication.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
