Performance evaluation of a new on-demand molecular test for the rapid identification of severe acute respiratory syndrome coronavirus 2 in pediatric and adult patients
- PMID: 36406396
- PMCID: PMC9670180
- DOI: 10.3389/fmicb.2022.999783
Performance evaluation of a new on-demand molecular test for the rapid identification of severe acute respiratory syndrome coronavirus 2 in pediatric and adult patients
Abstract
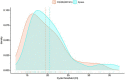
The rapid spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection has increased the need to identify additional rapid diagnostic tests for an accurate and early diagnosis of infection. Here, we evaluated the diagnostic performance of the cartridge-based reverse transcription polymerase chain reaction (RT-PCR) test STANDARD M10 SARS-CoV-2 (SD Biosensor Inc., Suwon, South Korea), targeting the ORF1ab and E gene of SARS-CoV-2, and which can process up to eight samples in parallel in 60 min. From January 2022 to March 2022, STANDARD™ M10 assay performance was compared with Xpert® Xpress SARS-CoV-2 (Cepheid, Sunnyvale CA) on 616 nasopharyngeal swabs from consecutive pediatric (N = 533) and adult (N = 83) patients presenting at the "Istituto di Ricovero e Cura a Carattere Scientifico" (IRCCS) Ospedale Pediatrico Bambino Gesù, Roma. The overall performance of STANDARD M10 SARS-CoV-2 was remarkably and consistently comparable to the Xpert® Xpress SARS-CoV-2 with an overall agreement of 98% (604/616 concordant results), and negligible differences in time-to-result (60 min vs. 50 min, respectively). When the Xpert® Xpress SARS-CoV-2 results were considered as the reference, STANDARD™ M10 SARS-CoV-2 had 96.5% sensitivity and 98.4% specificity. STANDARD M10 SARS-CoV-2 can thus be safely included in diagnostic pathways because it rapidly and accurately identifies SARS-CoV-2 present in nasopharyngeal swabs.
Keywords: COVID-19; RT-PCR testing; SARS-CoV-2; comparative diagnostic accuracy; molecular diagnosis.
Copyright © 2022 Colagrossi, Costabile, Scutari, Cento, Coltella, Reale, Scilipoti, Villani, Alteri, Perno and Russo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Alteri C., Cento V., Antonello M., Colagrossi L., Merli M., Ughi N., et al. (2020). Detection and quantification of SARS-CoV-2 by droplet digital PCR in real-time PCR negative nasopharyngeal swabs from suspected COVID-19 patients. PLoS One 15:e0236311. 10.1371/journal.pone.0236311 - DOI - PMC - PubMed
-
- Alteri C., Scutari R., Costabile V., Colagrossi L., La Rosa K. Y., Agolini E., et al. (2022). Epidemiological characterization of SARS-CoV-2 variants in children over the four COVID-19 waves and correlation with clinical presentation. Sci. Rep. 12:12814. 10.1038/s41598-022-17020-6 - DOI - PMC - PubMed
-
- Burkhard-Koren N. M., Haberecker M., Maccio U., Ruschitzka F., Schuepbach R. A., Zinkernagel A. S., et al. (2021). Higher prevalence of pulmonary macrothrombi in SARS-CoV-2 than in influenza A: Autopsy results from ‘Spanish flu’ 1918/1919 in Switzerland to Coronavirus disease 2019. J. Pathol. Clin. Res. 7 135–143. 10.1002/cjp2.189 - DOI - PMC - PubMed
-
- CDC (2022). Interim guidelines for collecting and handling of clinical specimens for COVID-19 testing. Available online at: https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specim... (accessed July, 2022).
LinkOut - more resources
Full Text Sources
Miscellaneous

