First molecular detection and genetic analysis of porcine circovirus 4 in the Southwest of China during 2021-2022
- PMID: 36406418
- PMCID: PMC9668871
- DOI: 10.3389/fmicb.2022.1052533
First molecular detection and genetic analysis of porcine circovirus 4 in the Southwest of China during 2021-2022
Abstract
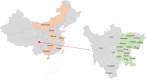
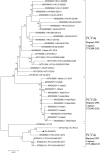
Porcine circovirus 4 (PCV4) was identified in 2019 as a novel circovirus species and then proved to be pathogenic to piglets. However, there is a lack of its prevalence in the Southwest of China. To investigate whether PCV4 DNA existed in the Southwest of China, 374 samples were collected from diseased pigs during 2021-2022 and detected by a real-time PCR assay. The results showed that the positive rate of PCV4 was 1.34% (5/374) at sample level, and PCV4 was detected in two of 12 cities, demonstrating that PCV4 could be detected in pig farms in the Southwest of China, but its prevalence was low. Furthermore, one PCV4 strain (SC-GA2022ABTC) was sequenced in this study and shared a high identity (98.1-99.7%) with reference strains at the genome level. Combining genetic evolution analysis with amino acid sequence analysis, three genotypes PCV4a, PCV4b, and PCV4c were temporarily identified, and the SC-GA2022ABTC strain belonged to PCV4c with a specific amino acid pattern (239V for Rep protein, 27N, 28R, and 212M for Cap protein). Phylogenetic tree and amino acid alignment showed that PCV4 had an ancient ancestor with mink circovirus. In conclusion, the present study was the first to report the discovery and the evolutionary analysis of the PCV4 genome in pig herds of the Southwest of China and provide insight into the molecular epidemiology of PCV4.
Keywords: genetic analysis; genotype; molecular detection; porcine circovirus 4; the Southwest of China.
Copyright © 2022 Xu, You, Wu, Zhu, Sun, Lai, Ai, Zhou and Xu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
First Molecular Detection and Genetic Analysis of a Novel Porcine Circovirus (Porcine Circovirus 4) in Dogs in the World.Microbiol Spectr. 2023 Feb 2;11(2):e0433322. doi: 10.1128/spectrum.04333-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36728419 Free PMC article.
-
Cross-species transmission of an emerging porcine circovirus (PCV4): First molecular detection and retrospective investigation in dairy cows.Vet Microbiol. 2022 Oct;273:109528. doi: 10.1016/j.vetmic.2022.109528. Epub 2022 Aug 5. Vet Microbiol. 2022. PMID: 35944390
-
Simultaneous detection and genetic characterization of porcine circovirus 2 and 4 in Henan province of China.Gene. 2022 Jan 15;808:145991. doi: 10.1016/j.gene.2021.145991. Epub 2021 Oct 6. Gene. 2022. PMID: 34626723
-
Current knowledge on epidemiology and evolution of novel porcine circovirus 4.Vet Res. 2022 May 31;53(1):38. doi: 10.1186/s13567-022-01053-w. Vet Res. 2022. PMID: 35642044 Free PMC article. Review.
-
Porcine circoviruses: current status, knowledge gaps and challenges.Virus Res. 2020 Sep;286:198044. doi: 10.1016/j.virusres.2020.198044. Epub 2020 Jun 2. Virus Res. 2020. PMID: 32502553 Review.
Cited by
-
Rescue and characterization of PCV4 infectious clones: pathogenesis and immune response in piglets.Front Microbiol. 2024 Jul 29;15:1443119. doi: 10.3389/fmicb.2024.1443119. eCollection 2024. Front Microbiol. 2024. PMID: 39135875 Free PMC article.
-
Prevalence and genetic characterization of porcine circovirus type 2, 3 and 4 in the upper Northern region of Thailand.Vet Res Commun. 2025 Jun 10;49(4):218. doi: 10.1007/s11259-025-10786-w. Vet Res Commun. 2025. PMID: 40493306 Free PMC article.
-
Development of a Triplex Real-Time PCR Method for the Simultaneous Detection of Porcine Circovirus 2, 3, and 4 in China Between 2023 and 2024.Viruses. 2025 May 29;17(6):777. doi: 10.3390/v17060777. Viruses. 2025. PMID: 40573369 Free PMC article.
-
Revisiting Porcine Circovirus Infection: Recent Insights and Its Significance in the Piggery Sector.Vaccines (Basel). 2023 Jul 31;11(8):1308. doi: 10.3390/vaccines11081308. Vaccines (Basel). 2023. PMID: 37631876 Free PMC article. Review.
-
Development of a duplex real-time recombinase aided amplification assay for the simultaneous and rapid detection of PCV3 and PCV4.Virol J. 2025 Feb 1;22(1):23. doi: 10.1186/s12985-025-02625-w. Virol J. 2025. PMID: 39893430 Free PMC article.
References
-
- Chen N., Xiao Y., Li X., Li S., Xie N., Yan X., et al. (2021). Development and application of a quadruplex real-time PCR assay for differential detection of porcine circoviruses (PCV1 to PCV4) in Jiangsu province of China from 2016 to 2020. Transbound. Emerg. Dis. 68 1615–1624. 10.1111/tbed.13833 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

