Antimicrobial resistance and genomic characterization of Salmonella enterica serovar Senftenberg isolates in production animals from the United States
- PMID: 36406424
- PMCID: PMC9668867
- DOI: 10.3389/fmicb.2022.979790
Antimicrobial resistance and genomic characterization of Salmonella enterica serovar Senftenberg isolates in production animals from the United States
Abstract
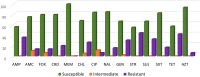
In the USA, Salmonella enterica subspecies enterica serovar Senftenberg is among the top five serovars isolated from food and the top 11 serovars isolated from clinically ill animals. Human infections are associated with exposure to farm environments or contaminated food. The objective of this study was to characterize S. Senftenberg isolates from production animals by analyzing phenotypic antimicrobial resistance profiles, genomic features and phylogeny. Salmonella Senftenberg isolates (n = 94) from 20 US states were selected from NVSL submissions (2014-2017), tested against 14 antimicrobial drugs, and resistance phenotypes determined. Resistance genotypes were determined using whole genome sequencing analysis with AMRFinder and the NCBI and ResFinder databases with ABRicate. Plasmids were detected using PlasmidFinder. Integrons were detected using IntFinder and manual alignment with reference genes. Multilocus-sequence-typing (MLST) was determined using ABRicate with PubMLST database, and phylogeny was determined using vSNP. Among 94 isolates, 60.6% were resistant to at least one antimicrobial and 39.4% showed multidrug resistance. The most prevalent resistance findings were for streptomycin (44.7%), tetracycline (42.6%), ampicillin (36.2%) and sulfisoxazole (32.9%). The most commonly found antimicrobial resistance genes were aac(6')-Iaa (100%), aph(3″)-Ib and aph(6)-Id (29.8%) for aminoglycosides, followed by bla TEM-1 (26.6%) for penicillins, sul1 (25.5%) and sul2 (23.4%) for sulfonamides and tetA (23.4%) for tetracyclines. Quinolone-resistant isolates presented mutations in gyrA and/or parC genes. Class 1 integrons were found in 37 isolates. Thirty-six plasmid types were identified among 77.7% of the isolates. Phylogenetic analysis identified two distinct lineages of S. Senftenberg that correlated with the MLST results. Isolates were classified into two distinct sequence types (ST): ST14 (97.9%) and ST 185 (2.1%). The diversity of this serotype suggests multiple introductions into animal populations from outside sources. This study provided antimicrobial susceptibility and genomic characteristics of S. Senftenberg clinical isolates from production animals in the USA during 2014 to 2017. This study will serve as a base for future studies focused on the phenotypic and molecular antimicrobial characterization of S. Senftenberg isolates in animals. Monitoring of antimicrobial resistance to detect emergence of multidrug-resistant strains is critical.
Keywords: Salmonella Senftenberg; antimicrobial resistance; phylogeny; resistance genes; whole genome sequencing.
Copyright © 2022 Srednik, Morningstar-Shaw, Hicks, Mackie and Schlater.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Antimicrobial resistance and genomic characterization of Salmonella Dublin isolates in cattle from the United States.PLoS One. 2021 Sep 21;16(9):e0249617. doi: 10.1371/journal.pone.0249617. eCollection 2021. PLoS One. 2021. PMID: 34547028 Free PMC article.
-
Whole-genome sequencing-based characterization of Salmonella enterica Serovar Enteritidis and Kentucky isolated from laying hens in northwest of Iran, 2022-2023.Gut Pathog. 2025 Jan 16;17(1):2. doi: 10.1186/s13099-025-00679-3. Gut Pathog. 2025. PMID: 39819347 Free PMC article.
-
Molecular mechanisms of resistance in multidrug-resistant serovars of Salmonella enterica isolated from foods in Germany.J Antimicrob Chemother. 2005 Dec;56(6):1025-33. doi: 10.1093/jac/dki365. Epub 2005 Oct 14. J Antimicrob Chemother. 2005. PMID: 16227350
-
Comparison of antimicrobial resistance among Salmonella enterica serovars isolated from Canadian turkey flocks, 2013 to 2021.Poult Sci. 2023 Jun;102(6):102655. doi: 10.1016/j.psj.2023.102655. Epub 2023 Mar 16. Poult Sci. 2023. PMID: 37030258 Free PMC article.
-
Genomic characterization of Salmonella enterica serovar Choleraesuis from Brazil reveals a swine gallbladder isolate harboring colistin resistance gene mcr-1.1.Braz J Microbiol. 2022 Dec;53(4):1799-1806. doi: 10.1007/s42770-022-00812-3. Epub 2022 Aug 19. Braz J Microbiol. 2022. PMID: 35984599 Free PMC article.
Cited by
-
Genomic insights into the serovar prevalence, antimicrobial resistance gene, and genetic diversity of Salmonella enterica in Mexico.PLoS One. 2025 May 15;20(5):e0323872. doi: 10.1371/journal.pone.0323872. eCollection 2025. PLoS One. 2025. PMID: 40373083 Free PMC article.
-
Phenotypic and WGS-derived antibiotic resistance patterns of Salmonella Enteritidis isolates from retail meat and environment during 2014 to 2019 in China.Front Microbiol. 2025 Jan 27;16:1502138. doi: 10.3389/fmicb.2025.1502138. eCollection 2025. Front Microbiol. 2025. PMID: 39931381 Free PMC article.
-
Exposing Salmonella Senftenberg and Escherichia coli Strains Isolated from Poultry Farms to Formaldehyde and Lingonberry Extract at Low Concentrations.Int J Mol Sci. 2023 Sep 26;24(19):14579. doi: 10.3390/ijms241914579. Int J Mol Sci. 2023. PMID: 37834022 Free PMC article.
-
vSNP: a SNP pipeline for the generation of transparent SNP matrices and phylogenetic trees from whole genome sequencing data sets.BMC Genomics. 2024 Jun 1;25(1):545. doi: 10.1186/s12864-024-10437-5. BMC Genomics. 2024. PMID: 38822271 Free PMC article.
-
Association between clinical-biological characteristics of Klebsiella pneumoniae and 28-day mortality in patients with bloodstream infection.BMC Microbiol. 2024 Dec 30;24(1):552. doi: 10.1186/s12866-024-03714-6. BMC Microbiol. 2024. PMID: 39736549 Free PMC article.
References
-
- Boumart Z., Roche S. M., Lalande F., Virlogeux-Payant I., Hennequet-Antier3 C., Menanteau P., et al. . (2012). Heterogeneity of Persistence of Salmonella enterica Serotype Senftenberg strains could explain the emergence of this serotype in poultry flocks. PLoS One 7:e35782. doi: 10.1371/journal.pone.0035782, PMID: - DOI - PMC - PubMed
-
- Carattoli A., Zankari E., García-Fernández A., Voldby Larsen M., Lund O., Villa L., et al. . (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–903. doi: 10.1128/AAC.02412-14, PMID: - DOI - PMC - PubMed
-
- Carroll L. M., Gaballa A., Guldimann C., Sullivan G., Henderson L. O., Wiedmann M. (2019). Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. MBio 10, e00853–e00819. doi: 10.1128/mBio.00853-19, PMID: - DOI - PMC - PubMed
-
- Center for Genomic Epidemiology (1998). Technical university of Denmark, Kongens. Lyngby, Denmark. https://cge.cbs.dtu.dk/services/IntFinder-1.0/ (Accessed April 2022).
LinkOut - more resources
Full Text Sources
Miscellaneous

