Biochemical, genomic and structural characteristics of the Acr3 pump in Exiguobacterium strains isolated from arsenic-rich Salar de Huasco sediments
- PMID: 36406427
- PMCID: PMC9671657
- DOI: 10.3389/fmicb.2022.1047283
Biochemical, genomic and structural characteristics of the Acr3 pump in Exiguobacterium strains isolated from arsenic-rich Salar de Huasco sediments
Abstract
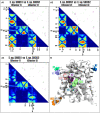
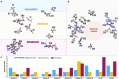
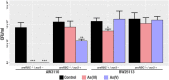
Arsenic is a highly toxic metalloid of major concern for public safety. However, microorganisms have several resistance mechanisms, particularly the expression of arsenic pumps is a critical component for bacterial ability to expel it and decrease intracellular toxicity. In this study, we aimed to characterize the biochemical, structural, and genomic characteristics of the Acr3 pump among a group of Exiguobacterium strains isolated from different sites of the arsenic-rich Salar de Huasco (SH) ecosystem. We also determined whether the differences in As(III) resistance levels presented by the strains could be attributed to changes in the sequence or structure of this protein. In this context, we found that based on acr3 sequences the strains isolated from the SH grouped together phylogenetically, even though clustering based on gene sequence identity did not reflect the strain's geographical origin. Furthermore, we determined the genetic context of the acr3 sequences and found that there are two versions of the organization of acr3 gene clusters, that do not reflect the strain's origin nor arsenic resistance level. We also contribute to the knowledge regarding structure of the Acr3 protein and its possible implications on the functionality of the pump, finding that although important and conserved components of this family of proteins are present, there are several changes in the amino acidic sequences that may affect the interactions among amino acids in the 3D model, which in fact are evidenced as changes in the structure and residues contacts. Finally, we demonstrated through heterologous expression that the Exiguobacterium Acr3 pump does indeed improve the organisms As resistance level, as evidenced in the complemented E. coli strains. The understanding of arsenic detoxification processes in prokaryotes has vast biotechnological potential and it can also provide a lot of information to understand the processes of evolutionary adaptation.
Keywords: ACR3; Exiguobacterium; arsenic; efflux pumps; resistance.
Copyright © 2022 Castro-Severyn, Pardo-Esté, Araya-Durán, Gariazzo, Cabezas, Valdés, Remonsellez and Saavedra.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Comparative Genomics Analysis of a New Exiguobacterium Strain from Salar de Huasco Reveals a Repertoire of Stress-Related Genes and Arsenic Resistance.Front Microbiol. 2017 Mar 21;8:456. doi: 10.3389/fmicb.2017.00456. eCollection 2017. Front Microbiol. 2017. PMID: 28377753 Free PMC article.
-
Genomic Variation and Arsenic Tolerance Emerged as Niche Specific Adaptations by Different Exiguobacterium Strains Isolated From the Extreme Salar de Huasco Environment in Chilean - Altiplano.Front Microbiol. 2020 Jul 15;11:1632. doi: 10.3389/fmicb.2020.01632. eCollection 2020. Front Microbiol. 2020. PMID: 32760381 Free PMC article.
-
Arsenic Response of Three Altiplanic Exiguobacterium Strains With Different Tolerance Levels Against the Metalloid Species: A Proteomics Study.Front Microbiol. 2019 Sep 26;10:2161. doi: 10.3389/fmicb.2019.02161. eCollection 2019. Front Microbiol. 2019. PMID: 31611848 Free PMC article.
-
Comparative Analysis of Arsenic Transport and Tolerance Mechanisms: Evolution from Prokaryote to Higher Plants.Cells. 2022 Sep 2;11(17):2741. doi: 10.3390/cells11172741. Cells. 2022. PMID: 36078150 Free PMC article. Review.
-
The Arsenic Detoxification System in Corynebacteria: Basis and Application for Bioremediation and Redox Control.Adv Appl Microbiol. 2017;99:103-137. doi: 10.1016/bs.aambs.2017.01.001. Epub 2017 Mar 6. Adv Appl Microbiol. 2017. PMID: 28438267 Review.
Cited by
-
Exploring the prokaryote-eukaryote interplay in microbial mats from an Andean athalassohaline wetland.Microbiol Spectr. 2024 Apr 2;12(4):e0007224. doi: 10.1128/spectrum.00072-24. Epub 2024 Mar 8. Microbiol Spectr. 2024. PMID: 38456669 Free PMC article.
-
Distinct bacterial community structures and arsenic biotransformation gene profiles in dust.Front Microbiol. 2025 Jul 30;16:1607082. doi: 10.3389/fmicb.2025.1607082. eCollection 2025. Front Microbiol. 2025. PMID: 40809051 Free PMC article.
-
Comparative genomic analysis and characterization of novel high-quality draft genomes from the coal metagenome.World J Microbiol Biotechnol. 2024 Nov 1;40(12):370. doi: 10.1007/s11274-024-04174-w. World J Microbiol Biotechnol. 2024. PMID: 39485561
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

